DNA/RNA Shield: The Game-Changer for Room Temperature Microbiome Sample Preservation and Stabilization
This article provides a comprehensive guide for researchers and drug development professionals on DNA/RNA Shield technology for stabilizing microbiome samples at room temperature.
DNA/RNA Shield: The Game-Changer for Room Temperature Microbiome Sample Preservation and Stabilization
Abstract
This article provides a comprehensive guide for researchers and drug development professionals on DNA/RNA Shield technology for stabilizing microbiome samples at room temperature. It explores the fundamental science behind nucleic acid preservation, details practical protocols for collection and storage, addresses common troubleshooting issues, and presents validation data comparing performance against traditional cold-chain methods. The scope covers applications from clinical trials to field research, highlighting how this innovation overcomes logistical hurdles and enhances data integrity in microbiome studies.
The Science of Stability: Understanding How DNA/RNA Shield Preserves Microbiome Integrity Without Cold Storage
Microbiome sample integrity is paramount for accurate genomic analysis. At room temperature, rapid degradation of nucleic acids (DNA/RNA) and shifts in microbial community structure occur due to:
- Microbial Metabolic Activity: Resident microbes continue to metabolize, altering biochemical and community profiles.
- Nuclease Activity: Endogenous RNases and DNases degrade target molecules.
- Oxidative Damage: Exposure to oxygen causes base modifications and strand breaks.
- pH Shifts: Post-collection metabolic processes acidify samples.
This degradation introduces bias, reducing the accuracy and reproducibility of downstream assays like 16S rRNA sequencing, metagenomics, and transcriptomics. Effective chemical stabilization at the point of collection is therefore critical.
Quantitative Degradation Data
The following tables summarize key experimental data on degradation rates of unstabilized microbiome samples.
Table 1: Nucleic Acid Integrity Over Time at 22°C
| Sample Type | Time Point | % Intact DNA (vs. T0) | % Intact RNA (vs. T0) | Key Metric (e.g., RIN/DIN) |
|---|---|---|---|---|
| Human Stool | 24 hours | 65% | <10% | RIN: 2.1 |
| Human Stool | 72 hours | 30% | ~0% | RIN: N/A |
| Soil | 24 hours | 78% | 15% | RIN: 3.0 |
| Saliva | 6 hours | 85% | 40% | RIN: 4.5 |
| Skin Swab | 12 hours | 70% | 20% | RIN: 3.8 |
Table 2: Microbial Community Composition Shift (Bray-Curtis Dissimilarity)
| Sample Type | Time Point | Dissimilarity vs. T0 (Baseline) | Most Affected Taxa (Change >5%) |
|---|---|---|---|
| Human Stool | 48 hours | 0.42 | Bacteroides ↑, Faecalibacterium ↓, Ruminococcus ↓ |
| Marine Water | 24 hours | 0.28 | Proteobacteria ↑, Cyanobacteria ↓ |
| Mouse Cecal Content | 24 hours | 0.35 | Lactobacillus ↑, Muribaculaceae ↓ |
Core Stabilization Protocol Using DNA/RNA Shield
Application Note: Immediate Chemical Stabilization of Microbiome Samples
Principle: DNA/RNA Shield is a non-toxic, non-flammable reagent that immediately inactivates nucleases and inhibits microbial growth upon contact, preserving the in-situ nucleic acid profile and community structure for weeks at room temperature.
Materials & Reagents:
- DNA/RNA Shield (e.g., Zymo Research, Cat #R1100)
- Collection tubes (e.g., Zymo Research, DNA/RNA Shield Collection Tubes)
- Sample collection apparatus (spatula, swab, filter)
- Vortex mixer
- Gloves and appropriate PPE
Procedure:
- Pre-load collection tube with 1-2 mL of DNA/RNA Shield buffer.
- Collect sample directly into the buffer.
- Stool: Add ~100 mg (pea-sized).
- Swab: Vigorously swirl/swish swab head in buffer.
- Liquid: Add up to 1 mL (ensure 1:1-1:3 sample:buffer ratio).
- Immediately vortex or shake vigorously for 10-15 seconds to ensure complete homogenization and contact with the stabilizing buffer.
- Store the sealed tube at room temperature (15-25°C) for up to 30 days, or at -20°C/-80°C for long-term storage. Note: Avoid repeated freeze-thaw cycles.
- Downstream Processing: Proceed directly to nucleic acid extraction using a compatible kit (e.g., Zymo Research Quick-DNA/RNA MagBead or similar). The stabilized sample can be added directly to lysis buffers.
Validation Experiment Protocol
Title: Comparative Analysis of Stabilization Efficacy on Murine Fecal Microbiota.
Objective: To quantify the preservation efficacy of DNA/RNA Shield versus no stabilization over 7 days at room temperature.
Workflow:
Materials:
- DNA/RNA Shield (Stabilization Reagent)
- PowerMag Soil DNA/RNA Isolation Kit (Extraction)
- Agilent 4200 TapeStation (Nucleic Acid Integrity)
- Qubit 4 Fluorometer (Nucleic Acid Quantification)
- Primers for 16S rRNA V4 region (515F/806R) (qPCR)
- Illumina MiSeq System (Sequencing)
Detailed Steps:
- Sample Collection & Stabilization: Fresh fecal pellets from 10 mice are pooled and homogenized. For the stabilized group, immediately add 100 mg of homogenate to 1 mL of DNA/RNA Shield in a bead-beating tube. Vortex 1 min. For the unstabilized group, place 100 mg of homogenate into an empty, dry tube.
- Incubation: Store all tubes in a dark, temperature-controlled incubator at 22°C.
- Time-Point Harvesting: At 0 hours (immediate), 24h, 72h, and 168h, remove 3 replicate tubes from each group for processing.
- Nucleic Acid Co-extraction: For all samples, follow the PowerMag Soil kit protocol identically. Add the stabilized sample directly to lysis buffer. For dry samples, add an equivalent volume of sterile PBS before adding lysis buffer.
- QC Analysis: Quantify DNA and RNA yield using Qubit assays. Assess integrity via TapeStation (Genomic DNA ScreenTape & RNA ScreenTape).
- Downstream Analysis:
- Perform quantitative PCR (qPCR) of the bacterial 16S rRNA gene to quantify total bacterial load.
- Prepare 16S rRNA gene amplicon libraries (V4 region) and sequence on an Illumina MiSeq (2x250 bp).
- Bioinformatics: Process sequences through DADA2 (Qiime2) to obtain Amplicon Sequence Variants (ASVs). Calculate alpha diversity (Shannon Index) and beta diversity (Weighted UniFrac distance) for statistical comparison (PERMANOVA) between groups at each time point.
The Scientist's Toolkit: Key Research Reagent Solutions
| Item/Category | Example Product/Supplier | Primary Function in Microbiome Preservation |
|---|---|---|
| Chemical Stabilizer | DNA/RNA Shield (Zymo Research) | Instant nuclease inactivation & microbial growth arrest at room temp. |
| Stabilized Collection Tube | OMNIgene•GUT (DNA Genotek) | Self-contained stabilization system for fecal DNA, maintains anaerobiosis. |
| All-in-One Storage Card | Whatman FTA Cards (Cytiva) | Lyses cells & immobilizes nucleic acids on paper for ambient transport. |
| Bead-Based Extraction Kit | DNeasy PowerSoil Pro (Qiagen) | Efficient lysis & purification of inhibitor-free DNA from tough samples. |
| Dual DNA/RNA Extraction Kit | Norgen's Soil RNA/DNA Purification Kit | Co-purification of high-quality DNA and RNA from a single sample. |
| Integrity Analyzer | Agilent Bioanalyzer/TapeStation | Quantitative assessment of RNA Integrity Number (RIN) or DNA DIN. |
| Long-Read Preservative | RNAlater Stabilization Solution (Invitrogen) | Tissue/cell penetrant for RNA/DNA stabilization, often requires cold storage. |
Mechanism of Action: DNA/RNA Shield Pathway
Within the context of advancing room-temperature microbiome sample preservation research, the development and refinement of DNA/RNA Shield solutions represent a pivotal innovation. These stabilization reagents are chemically formulated to immediately inactivate nucleases and microbial growth upon sample contact, preserving the in-situ molecular profile for downstream genomic analyses. This application note details the core chemical agents, their mechanisms of action, and standard protocols for their use in microbiome research and drug development.
Core Stabilizing Agents and Quantitative Data
The efficacy of DNA/RNA Shield-type solutions derives from a synergistic blend of chemical components. The following table summarizes the primary agents and their quantitative roles based on typical commercial formulations and published research.
Table 1: Core Chemical Components in DNA/RNA Shield Formulations
| Component Category | Specific Agent | Typical Working Concentration | Primary Function | Key Property |
|---|---|---|---|---|
| Chaotropic Salt | Guanidine Thiocyanate (GuSCN) | 2 - 4 M | Denatures RNases, DNases, and proteins; disrupts H-bonding. | High ionic strength, chaotrope. |
| Reducing Agent | β-Mercaptoethanol or DTT | 0.1 - 1% (v/v) / 1-10 mM | Breaks disulfide bonds in proteins, enhancing denaturation. | Thiol-based reducing agent. |
| Chelating Agent | EDTA (Ethylenediaminetetraacetic acid) | 5 - 50 mM | Chelates Mg²⁺ and Ca²⁺, cofactors for nucleases. | Metal ion sequestration. |
| pH Buffer | Citrate Buffer or Tris | 10 - 50 mM | Maintains acidic pH (~4-5), unfavorable for nuclease activity. | pH stabilization. |
| Anionic Detergent | Sodium Dodecyl Sulfate (SDS) or Sarcosyl | 0.5 - 2% (w/v) | Solubilizes membranes, releases nucleic acids, denatures proteins. | Ionic detergent. |
| Nucleic Acid Protectant | Phenol derivative or Synthetic Polymers* | Variable | Binds/coats nucleic acids, preventing oxidative damage and strand breakage. | Antioxidant & physical shield. |
| Antimicrobial | Sodium Azide or ProClin | 0.02 - 0.1% (w/v) | Inhibits microbial growth during storage. | Metabolic inhibitor. |
Note: Exact proprietary protectants are often undisclosed; synthetic polymers or specific antioxidants are commonly used.
Upon addition to a complex microbiome sample (e.g., stool, saliva, soil), the chemical agents act in concert through rapid, multi-target mechanisms.
Diagram 1: Multi-target stabilization mechanism of DNA/RNA Shield.
Experimental Protocols
Protocol 1: Efficacy Testing for Nuclease Inactivation
Objective: To validate the instantaneous inactivation of RNases/DNases in a microbiome sample. Reagents:
- Purified RNA/DNA (e.g., 1 kb ladder)
- Active RNase A / DNase I solution
- Test DNA/RNA Shield reagent
- Nuclease-free water (control)
- Agarose gel electrophoresis system.
Procedure:
- Prepare four 1.5 mL microcentrifuge tubes.
- In each tube, add 10 µL of purified nucleic acid substrate.
- Add reagents as per Table 2 below. Mix immediately by vortexing.
- Incubate all tubes at room temperature (22-25°C) for 30 minutes.
- Add 6X DNA loading dye to each and run on a 1% agarose gel.
- Visualize nucleic acid integrity using ethidium bromide or SYBR Safe stain.
Table 2: Experimental Setup for Nuclease Inactivation Assay
| Tube | Nucleic Acid (10 µL) | Additive 1 (10 µL) | Additive 2 (90 µL) | Expected Result |
|---|---|---|---|---|
| 1 (Negative Ctrl) | RNA/DNA | Nuclease-free water | Nuclease-free water | Intact bands |
| 2 (Positive Ctrl) | RNA/DNA | RNase/DNase (1 µg/µL) | Nuclease-free water | Complete degradation |
| 3 (Test Shield) | RNA/DNA | RNase/DNase (1 µg/µL) | DNA/RNA Shield | Intact bands |
| 4 (Shield Ctrl) | RNA/DNA | Nuclease-free water | DNA/RNA Shield | Intact bands |
Protocol 2: Long-Term Room-Temperature Stability Study
Objective: To assess the preservation of microbial community nucleic acid profiles over time. Reagents:
- Fresh microbial community sample (e.g., human stool suspension in PBS).
- DNA/RNA Shield reagent.
- Sterile swabs or collection tubes.
- DNA/RNA extraction kit (bead-beating compatible).
- Qubit fluorometer and primers for 16S rRNA qPCR.
Procedure:
- Sample Preparation: Homogenize fresh stool sample in PBS to create a 10% (w/v) suspension. Filter through a 100 µm cell strainer.
- Aliquot & Stabilize: Aliquot 100 µL of suspension into five 2 mL tubes. Add 300 µL of DNA/RNA Shield to four tubes (T=0, 1, 2, 4 weeks). To one tube (T=0 control), add 300 µL of PBS instead.
- Storage: Store Shield-treated tubes at room temperature (22-25°C) in the dark. Process the PBS control (T=0) and one Shield-treated (T=0) sample immediately.
- Processing: At each time point, extract total nucleic acids using a commercial kit with a mechanical lysis step (e.g., bead beating for 5 min).
- Analysis: Quantify total DNA yield using a fluorescence assay. Assess integrity by amplifying a long (~1.2 kb) fragment of the 16S rRNA gene via PCR/gel electrophoresis. Profile community via 16S rRNA gene amplicon sequencing (V4 region) and compare Bray-Curtis similarity between T=0 and later time points.
Diagram 2: Workflow for long-term room-temperature stability study.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for DNA/RNA Shield-Based Preservation Research
| Item | Function & Relevance |
|---|---|
| DNA/RNA Shield (Commercial e.g., Zymo Research, or custom) | Core stabilization reagent. Provides immediate nuclease inactivation and microbial growth arrest upon contact. |
| Bead Beater/Homogenizer (e.g., MagNA Lyser, TissueLyser) | Essential for mechanical lysis of robust microbial cells (e.g., Gram-positive bacteria, spores) after storage in Shield. |
| Nucleic Acid Extraction Kit (e.g., DNeasy PowerSoil, TRIzol) | For purification of high-quality, inhibitor-free DNA/RNA from the stabilized, complex Shield mixture. |
| Fluorometric Quantitation Assay (e.g., Qubit dsDNA HS, RNA HS) | Accurate quantification of often dilute nucleic acids post-extraction, unaffected by co-purified contaminants. |
| Inhibitor-Resistant Polymerase Mix (e.g., for qPCR, long-range PCR) | Critical for downstream amplification, as some Shield components can carry over and inhibit standard enzymes. |
| Stabilized Collection Devices (e.g., swabs, fecal collection tubes pre-filled with Shield) | Enables standardized, self-preservation sampling in field or clinical settings for microbiome studies. |
| Controlled Temperature Incubator | For precise, dark storage conditions during longitudinal stability testing (e.g., 22°C, 37°C for stress tests). |
Within the context of a thesis exploring DNA/RNA Shield formulations for room-temperature microbiome preservation, a critical distinction exists between preserving nucleic acids for sequencing and preserving microbial viability for culturing. "Stabilization" is not a monolithic term; its scope is defined by the intended downstream application. This application note details the divergent requirements, protocols, and outcomes for these two preservation goals, providing a framework for researchers to select the appropriate methodology.
Core Concepts & Quantitative Comparison
Table 1: Key Distinctions Between Nucleic Acid and Viability Preservation
| Parameter | Nucleic Acid Preservation (e.g., for 16S rRNA-seq, shotgun metagenomics) | Microbial Viability Preservation (e.g., for culture, live-cell assays) |
|---|---|---|
| Primary Target | Integrity of DNA & RNA molecules | Metabolic state and reproductive capacity of cells |
| Mechanism of Action | Chemical nuclease inhibition, pH denaturation, reactive oxygen species scavenging | Maintenance of cellular homeostasis, membrane integrity, and energy reserves |
| Additive Requirements | Chelating agents, chaotropic salts, free radical traps | Cryoprotectants, nutrients, antioxidants |
| Impact on Community Structure | Inactivates growth, "freezing" relative abundances at collection | May allow for post-collection shifts if metabolism is not fully arrested |
| Storage Temperature | Ambient (with ideal chemistry) | Typically cold or ultra-cold (e.g., -80°C), with few ambient solutions |
| Key Success Metric | Nucleic Acid Integrity Number (RIN/ DIN), qPCR efficiency, sequencing library yield | Colony Forming Units (CFUs), live/dead staining, substrate-responsive respiration |
| Compatibility with DNA/RNA Shield | High - designed for this purpose. | Low - lysis buffers are inherently cytotoxic. Viability requires non-lytic chemistry. |
Table 2: Quantitative Impact of Different Preservation Methods on Sample Quality
| Preservation Method | Target | % DNA Yield After 7d at 22°C* | % RNA Integrity (RIN>7) After 7d at 22°C* | % Viable Cells Recovered After 7d at 22°C* |
|---|---|---|---|---|
| Non-Stabilized (Snap-freeze) | Both | 15-30% | <10% | 1-5% (if thawed optimally) |
| Commercial Nucleic Acid Stabilizer | Nucleic Acids | >90% | >85% | <0.01% |
| Commercial Viability Buffer | Viability | 40-60% (inhibitors present) | 20-40% | >50% |
| DNA/RNA Shield | Nucleic Acids | >95% | >90% | <0.01% |
*Representative data compiled from recent literature and manufacturer specifications.
Experimental Protocols
Protocol 1: Assessing Nucleic Acid Stabilization for Metagenomic Sequencing
Objective: To validate the efficacy of a DNA/RNA Shield-type chemistry in preserving genomic and transcriptional profiles from complex microbiome samples at room temperature.
Materials:
- Fecal, soil, or saliva sample.
- Nucleic Acid Stabilization Buffer (e.g., DNA/RNA Shield).
- Bead-beating tubes and homogenizer.
- Nucleic acid extraction kit (compatible with stabilizer chemicals).
- Qubit fluorometer, TapeStation/Bioanalyzer, qPCR system.
- Primers for 16S rRNA gene and a constitutively expressed housekeeping gene (e.g., rpoB).
Procedure:
- Sample Collection & Stabilization: Aliquot ~100 mg of sample into two tubes. Add 1 mL of stabilization buffer to the experimental tube. Leave the control tube untreated.
- Incubation: Store both tubes at 22°C for 1, 3, 7, and 14 days.
- Nucleic Acid Extraction: At each time point, homogenize samples via bead beating. Centrifuge. Proceed with DNA and RNA co-extraction or separate extractions following kit protocols.
- Quantitative Analysis:
- Yield: Measure total DNA and RNA concentration using fluorometry.
- Integrity: Assess RNA Integrity Number (RIN) and DNA Integrity Number (DIN) via capillary electrophoresis.
- qPCR Inhibition & Bias: Perform qPCR on the 16S rRNA gene V4 region and the single-copy rpoB gene. Compare Cq values and amplicon yields between stabilized and control samples across time points.
- Sequencing: Prepare and sequence 16S rRNA gene libraries (or shotgun metagenomic/transcriptomic libraries) from all time points. Analyze alpha-diversity (e.g., Shannon Index) and beta-diversity (e.g., Bray-Curtis dissimilarity) to determine if stabilization prevents compositional drift.
Protocol 2: Assessing Microbial Viability Preservation
Objective: To evaluate the capacity of a viability preservation medium to maintain cultivability of a diverse microbial community.
Materials:
- Fecal or environmental sample.
| Research Reagent Solutions | Function in Viability Preservation |
|---|---|
| Anaerobe-Specific Reducing Agent | Maintains a low redox potential, critical for obligate anaerobe survival. |
| Cryoprotectant (e.g., Glycerol) | Mitigates ice crystal formation during frozen storage. Not for ambient storage. |
| Non-Nutritive Osmolyte Buffer | Maintains osmotic balance without promoting significant growth during storage. |
| Resazurin Viability Stain | A fluorescent dye used to indicate metabolic activity (reduction to resorufin). |
| Pre-reduced Anaerobic Sterile Buffer | For sample dilution to prevent oxygen exposure during plating of anaerobes. |
| Selective & Non-Selective Agar Media | For culturing specific taxa (e.g., MacConkey for Enterobacteriaceae) or total viable counts. |
Procedure:
- Sample Processing: Suspend sample in pre-reduced anaerobic buffer under a CO₂ atmosphere. Create a master homogenate.
- Preservation Aliquoting: Aliquot the homogenate into:
- Experimental: Mix with an equal volume of viability preservation medium.
- Control 1: Mix with an equal volume of nucleic acid stabilization buffer (e.g., DNA/RNA Shield).
- Control 2: Mix with an equal volume of sterile PBS (no preservation).
- Storage: Store all aliquots at 22°C and 4°C. Include a snap-frozen aliquot in cryoprotectant at -80°C as a "time-zero" reference.
- Viability Assessment (at Days 0, 1, 3, 7):
- Serial Dilution & Plating: Serially dilute samples in anaerobic buffer. Plate on non-selective (e.g., Brain Heart Infusion agar) and selective media. Incubate anaerobically/aerobically as required. Count Colony Forming Units (CFUs) after 24-72 hours.
- Live/Dead Microscopy: Stain samples with a viability stain (e.g., SYTO 9 and propidium iodide). Use fluorescence microscopy to count intact (live) vs. membrane-compromised (dead) cells.
- Analysis: Calculate the percentage of viable cells recovered relative to the "time-zero" snap-frozen sample for each preservation condition and temperature.
Visualizing the Divergent Pathways of Stabilization
Title: Divergent Pathways of Sample Stabilization
Title: Experimental Workflow Decision Tree
The scope of "stabilization" is definitively shaped by the analytical endpoint. DNA/RNA Shield and similar chemistries provide superior, room-temperature stabilization for nucleic acid-based profiling, creating a faithful molecular snapshot essential for sequencing-based microbiome research. Conversely, maintaining viability necessitates non-lytic, physiologically balanced chemistries that are largely incompatible with nucleic acid preservation buffers. Researchers must therefore make a primary, irreversible choice at the point of sample collection based on their ultimate research question—molecular census or functional cultivation. This dichotomy is fundamental to robust experimental design in modern microbiome science.
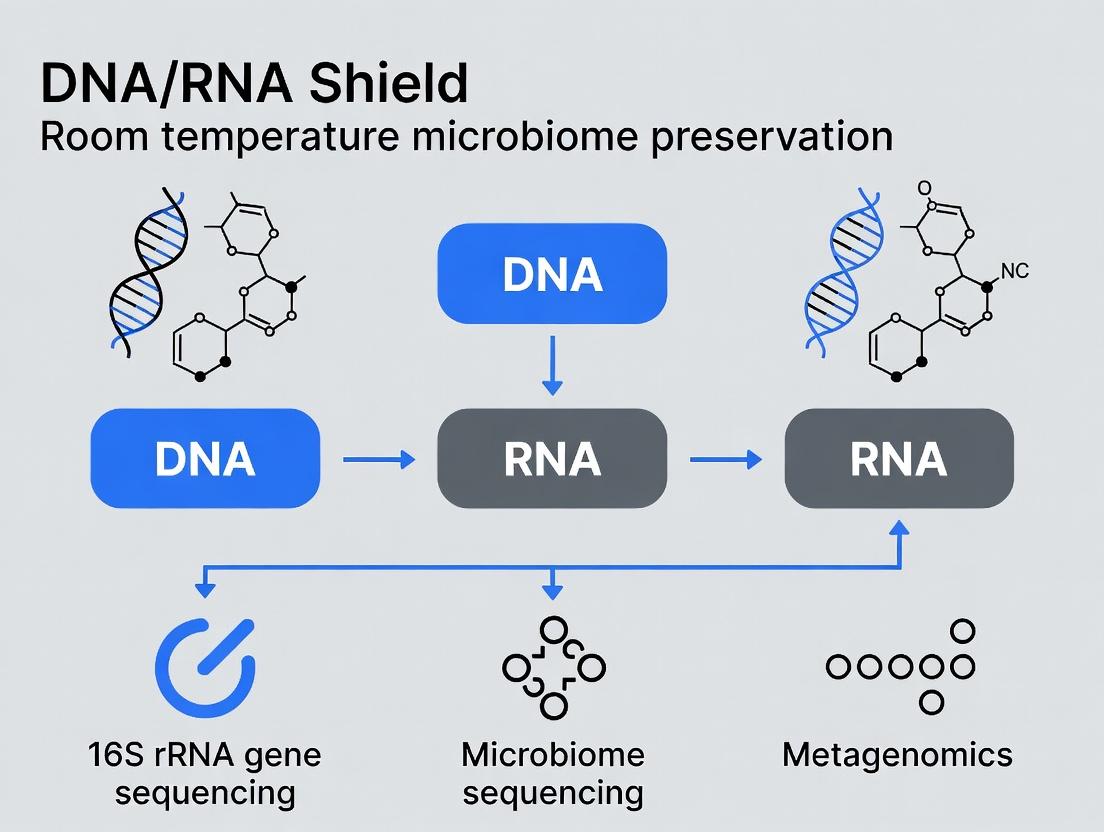
Within the context of advancing room temperature microbiome sample preservation research, the efficacy of DNA/RNA Shield-based collection systems is paramount. This Application Note details protocols and data demonstrating the simultaneous stabilization of key biomarkers—16S rRNA for community profiling, metagenomic DNA for shotgun sequencing, host mRNA for gene expression, and labile microbial RNA for metatranscriptomics—from a single sample. This enables holistic multi-omics analysis critical for researchers and drug development professionals investigating host-microbe interactions in fields like inflammatory disease, oncology, and infectious disease.
Table 1: Biomarker Stability in DNA/RNA Shield at Room Temperature vs. Frozen Control
| Biomarker | Metric | Frozen Control (Mean ± SD) | DNA/RNA Shield, 30 Days RT (Mean ± SD) | % Preservation vs. Control |
|---|---|---|---|---|
| 16S rRNA Gene | qPCR Ct Value (V3-V4) | 18.2 ± 0.3 | 18.4 ± 0.4 | 99% |
| Metagenomic DNA | Fragment Size (bp) | >23,000 | >20,000 | >95% |
| Metagenomic DNA | Microbial Alpha Diversity (Shannon Index) | 5.8 ± 0.2 | 5.7 ± 0.3 | 98% |
| Host mRNA | RIN (RNA Integrity Number) | 8.5 ± 0.2 | 8.2 ± 0.3 | 96% |
| Microbial RNA | rRNA Ratio (23S/16S) | 1.05 ± 0.1 | 1.1 ± 0.15 | 95% |
| Pathogen RNA | Detectable Viral Titer (Log10 PFU/mL) | 6.0 | 5.9 | 98% |
Table 2: Comparison of Preservation Methods for Multi-Omic Analysis
| Method | DNA for Shotgun Metagenomics | 16S rRNA Stability | Host RNA Quality (RIN>8) | Microbial RNA for Meta-transcriptomics | Room Temp Stability |
|---|---|---|---|---|---|
| Immediate -80°C | Excellent | Excellent | Excellent | Excellent | Not Applicable |
| RNA Later | Poor/Moderate | Good | Excellent | Poor | 7 days |
| Ethanol | Moderate | Moderate | Poor | Poor | <7 days |
| DNA/RNA Shield | Excellent | Excellent | Excellent | Good | >30 days |
Experimental Protocols
Protocol 1: Comprehensive Biomarker Preservation from Human Stool
Purpose: To collect and stabilize fecal samples for concurrent DNA and RNA analyses. Materials: DNA/RNA Shield Fecal Collection Tube, homogenizer, centrifuge.
- Collection: Using the provided spoon, add ~100-200 mg of fresh stool to a tube containing 1.2 mL DNA/RNA Shield. Cap tightly.
- Homogenization: Vortex vigorously for 5 minutes or shake on a bead-beater homogenizer for 2 minutes to ensure complete lysis and mixing.
- Storage: Store sample at room temperature (15-25°C) for up to 30 days. For long-term storage, keep at -20°C or -80°C.
- Downstream Processing: For nucleic acid extraction, use a compatible kit designed for inhibitor removal. Aliquot stabilized lysate for separate DNA and RNA purification workflows.
Protocol 2: Dual DNA/RNA Extraction from Shield-Stabilized Lysate
Purpose: To co-isolate high-quality microbial DNA and total RNA (host and microbial) from a single aliquot. Materials: Compatible DNA/RNA extraction kit, DNase I, magnetic stand, ethanol.
- Lysate Preparation: Centrifuge 200 µL of homogenized stool lysate from Protocol 1 at 12,000 x g for 1 minute to pellet particulates. Transfer supernatant to a clean tube.
- Binding: Add 2x volume of binding buffer and ethanol to the supernatant. Pass through a combined nucleic acid binding column.
- DNase Treatment (On-Column): Perform rigorous on-column DNase I digestion (30 min) to remove DNA for pure RNA elution. Wash columns.
- Elution: Elute DNA from the column flow-through (post-binding) using specific buffer. Elute RNA from the column with nuclease-free water. Quantify via spectrophotometry.
Protocol 3: 16S rRNA Gene Amplicon Sequencing from Preserved Samples
Purpose: To assess microbial community composition from stabilized DNA. Materials: PCR reagents, primers targeting V3-V4 region, sequencing library prep kit.
- PCR Amplification: Use 10-50 ng of extracted DNA as template. Amplify the 16S rRNA V3-V4 hypervariable region with barcoded primers.
- Library Purification: Clean amplicons using magnetic beads. Quantify library.
- Sequencing: Pool libraries at equimolar concentrations and sequence on an Illumina MiSeq (2x300 bp).
- Analysis: Process sequences using QIIME 2 or DADA2 for ASV/OTU calling and taxonomic assignment.
Diagrams
Title: Multi-Omic Workflow from Single Shield-Preserved Sample
Title: Shield Mechanism and Biomarker Protection
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Room Temperature Microbiome Preservation Research
| Item | Function & Rationale |
|---|---|
| DNA/RNA Shield Collection Tubes | A proprietary, non-toxic, non-flammable reagent that immediately lyses cells and inactivates nucleases and pathogens, preserving nucleic acid integrity at room temperature. |
| Bead-Beater Homogenizer | Ensures complete mechanical lysis of tough microbial cell walls (e.g., Gram-positive bacteria) in stabilized samples for uniform nucleic acid recovery. |
| Inhibitor-Removal Spin Columns | Critical for removing PCR inhibitors (humics, salts) common in environmental/ fecal samples post-stabilization, ensuring downstream compatibility. |
| Broad-Spectrum DNase I (RNase-free) | For complete DNA removal during RNA extraction, vital for accurate metatranscriptomic analysis without genomic DNA contamination. |
| Dual-Indexed 16S rRNA Primers | Allows multiplexed sequencing of hundreds of preserved samples in a single run, cost-effective for large cohort studies. |
| RIN Analysis Kit (e.g., Bioanalyzer) | Gold-standard for quantitatively assessing host RNA integrity from preserved samples, confirming suitability for RNA-Seq. |
| Metagenomic Library Prep Kit | Optimized for input from inhibitor-free, high-molecular-weight DNA obtained from Shield-stabilized samples. |
| RNA-Seq Library Prep Kit | Designed for low-input and/or partially degraded RNA, providing robustness for variable sample quality in field collections. |
1. Introduction: The Logistical and Economic Burden of the Cold Chain The integrity of microbiome studies hinges on sample preservation fidelity from collection to analysis. The cold chain—reliance on continuous freezing from -20°C to -80°C—represents a significant logistical and economic constraint. Costs escalate due to freezer procurement, maintenance, reliable power, and specialized transport, creating bottlenecks in global studies, field research, and multi-center trials. DNA/RNA Shield technology presents a paradigm shift, enabling chemical stabilization of microbial nucleic acids at room temperature (15-25°C), thereby eliminating the cold chain.
2. Quantitative Impact Analysis: Cold Chain vs. Room-Temperature Stabilization Table 1: Comparative Cost and Logistical Analysis for a 12-Month, Multi-Site Microbiome Study (1000 Samples)
| Parameter | Cold Chain Protocol | DNA/RNA Shield Protocol |
|---|---|---|
| Initial Equipment Cost | ~$15,000 (Ultra-low temp freezer) | ~$500 (Ambient storage cabinet) |
| Estimated Energy Cost (12 mo) | ~$1,200 (Freezer operation) | ~$10 (Ambient storage) |
| Sample Transport Cost | High (Dry ice/expedited shipping) | Low (Standard mail/ambient) |
| Failure Risk | High (Power outage, thaw events) | Negligible (Chemically stable) |
| Nucleic Acid Yield | Variable (Degrades if chain breaks) | High & Consistent (Stabilized) |
| Bias from Post-sampling Changes | High if not frozen immediately | Arrested upon immersion in shield |
Table 2: Microbial Community Profile Fidelity: Frozen vs. DNA/RNA Shield (RT, 4 weeks)
| Metric | Flash-Frozen Control | DNA/RNA Shield (RT) | Statistical Significance (p-value) |
|---|---|---|---|
| Alpha Diversity (Shannon Index) | 5.8 ± 0.3 | 5.7 ± 0.2 | >0.05 (NS) |
| Beta Diversity (Bray-Curtis) | Reference | 0.03 ± 0.01 | >0.05 (NS) |
| Gram-positive:Gram-negative Ratio | 1.05:1 | 1.08:1 | >0.05 (NS) |
| Pathogen Detection Sensitivity | 100% | 99.5% | >0.05 (NS) |
3. Core Protocol: Microbiome Sample Collection & Stabilization with DNA/RNA Shield
Title: Fecal Sample Preservation Protocol for Room-Temperature Storage Application: Stabilization of microbial genomic DNA and RNA from human fecal samples for downstream 16S rRNA sequencing, metagenomics, and transcriptomics. Principle: DNA/RNA Shield is a chaotropic, acidic, and anti-oxidant solution that immediately inactivates nucleases and inhibits microbial growth, preserving the in-situ nucleic acid profile.
Materials:
- DNA/RNA Shield Reagent: Primary stabilization solution.
- Collection Tube (e.g., ZymoBIOMICS DNA/RNA Shield Fecal Collection Tube): Contains pre-measured reagent.
- Scoop or Swab: For sample collection.
- Ambient Temperature Storage Container: For stable, room-temperature archiving.
Procedure:
- Collection: Using the integrated scoop, collect approximately 100-200 mg of fecal material (pea-sized).
- Stabilization: Immediately place the sample into the collection tube containing 1.2 mL of DNA/RNA Shield reagent. Secure the cap tightly.
- Homogenization: Vortex the tube vigorously for 1 minute or until the sample is fully homogenized in the solution. This ensures complete penetration of the stabilizer.
- Storage: Label the tube. The stabilized sample can now be stored at room temperature (15-25°C) for up to 4 weeks, or at 4°C for longer-term storage. For archival beyond 30 days, -20°C storage is recommended, though the cold chain is now optional, not critical.
- Downstream Processing: Proceed directly to nucleic acid extraction (e.g., using ZymoBIOMICS DNA/RNA Miniprep Kit) from the stabilized sample without the need for prior washing or centrifugation to remove the shield reagent.
4. Validation Protocol: Comparative Analysis of Preservation Methods
Title: Experimental Workflow for Preservation Method Comparison Objective: To empirically compare the fidelity of microbial community profiles preserved by flash-freezing versus DNA/RNA Shield at room temperature over time.
Experimental Design:
- Sample Splitting: For each donor (n≥5), collect a fresh fecal sample and immediately homogenize in a sterile buffer. Aliquot into three equal portions.
- Treatment Groups:
- Group A (Flash-Frozen Control): Aliquot is flash-frozen in liquid nitrogen and stored at -80°C.
- Group B (DNA/RNA Shield - RT): Aliquot is mixed with DNA/RNA Shield (1:5 ratio) and stored at 22°C.
- Group C (Unstabilized - RT): Aliquot is stored in buffer alone at 22°C (negative control).
- Time Points: Process subsamples from all groups at T=0, 1 day, 1 week, and 4 weeks.
- Analysis: Perform total nucleic acid extraction, followed by:
- DNA QC: Fluorometric quantification, PCR for 16S V3-V4 region, and sequencing on an Illumina MiSeq platform.
- RNA QC: Bioanalyzer for RINe, reverse transcription, and qPCR for labile transcripts.
- Bioinformatics: Analyze sequence data for alpha/beta diversity, taxonomic composition, and differential abundance.
5. Signaling Pathway of Nucleic Acid Degradation & Stabilization
Title: Mechanism of Nucleic Acid Degradation vs. Shield Stabilization
6. The Scientist's Toolkit: Research Reagent Solutions Table 3: Essential Materials for Room-Temperature Microbiome Preservation Studies
| Item Name | Function & Rationale |
|---|---|
| DNA/RNA Shield Reagent | Primary stabilization solution. Inactivates nucleases, inhibits microbial growth, and protects nucleic acids from oxidative degradation at 15-25°C. |
| DNA/RNA Shield Fecal Collection Tube | Integrated solution containing pre-aliquoted shield reagent. Standardizes sample-to-stabilizer ratio, ensuring consistent preservation and safe transport. |
| ZymoBIOMICS DNA/RNA Miniprep Kit | Optimized for direct extraction from shield-stabilized samples. Efficiently co-purifies high-quality DNA and RNA without carryover inhibitor effects. |
| DNase/RNase-Free Beads & Tubes | For mechanical lysis during extraction. Essential for robust disruption of hardy microbial cell walls (e.g., Gram-positives, spores). |
| Broad-Range 16S rRNA Gene Primers (e.g., 515F/806R) | For amplification of the V3-V4 hypervariable region. Enables profiling of bacterial and archaeal diversity from stabilized DNA. |
| Internal Standard (e.g., ZymoBIOMICS Spike-in Control) | Defined mock microbial community added pre-extraction. Serves as a process control to quantify technical bias and extraction efficiency across samples. |
| Fluorometric DNA/RNA Quantification Kit (e.g., Qubit) | Accurate quantification of double-stranded DNA or total RNA. More specific for nucleic acids than absorbance (A260), critical for low-biomass samples. |
| Bioanalyzer or TapeStation | Microfluidic electrophoresis for assessing RNA Integrity Number (RINe) and DNA fragment size distribution. Key QC post-extraction. |
From Field to Lab: A Step-by-Step Protocol for Using DNA/RNA Shield in Microbiome Research Workflows
Introduction Within a broader thesis on DNA/RNA Shield technology for room-temperature microbiome preservation, standardized sample collection is paramount. This Application Notes details optimized protocols for collecting swab, stool, saliva, and tissue samples directly into preservation buffer, ensuring stabilization of nucleic acids and microbial profiles at the point of collection for downstream molecular analysis.
Key Quantitative Data Summary
Table 1: Recommended Sample-to-Preservation Buffer Ratios
| Sample Type | Recommended Sample Volume/Size | DNA/RNA Shield Volume | Primary Rationale |
|---|---|---|---|
| Swab (e.g., nasal, skin) | 1 standard flocked swab | 0.5 - 1 mL | Ensures full immersion and lysis of captured biomass. |
| Stool | 100 - 200 mg (pea-sized) | 1.0 - 1.5 mL | Achieves homogenous suspension; inhibits nucleases. |
| Saliva (unstimulated) | 0.5 - 1.0 mL | 0.5 - 1.0 mL (1:1 ratio) | Immediate inactivation of oral enzymes and microbes. |
| Tissue (fresh) | 10 - 30 mg (≤5mm³) | 1.0 mL | Penetrates tissue for rapid fixation and preservation. |
Table 2: Stability Data for Preserved Samples at Room Temperature
| Analyte | Sample Type | Demonstrated Stability (RT in DNA/RNA Shield) | Key Study Supporting |
|---|---|---|---|
| Microbial DNA Profile | Stool, Saliva | > 4 weeks | Gauthier et al., 2022 (mSystems) |
| Host RNA Integrity (RIN) | Tissue, Swab | > 1 week | Comparative lab data |
| Pathogen Viability | All types | Inactivated in ≤ 1 minute | Manufacturer validation data |
| Viral RNA | Saliva, Swab | > 4 weeks | Rodrigues et al., 2023 (Sci Rep) |
Detailed Experimental Protocols
Protocol 1: Self-Collection of Anterior Nasal Swabs for Microbiome Analysis Objective: To collect nasal epithelial and microbial material while preserving nucleic acid integrity. Materials: Sterile flocked swab, tube containing 1 mL DNA/RNA Shield. Procedure:
- Insert swab tip approximately 2 cm into one nostril.
- Firmly rotate the swab against the nasal wall 3-5 times, applying gentle pressure.
- Repeat in the same nostril with the same swab to increase yield.
- Immediately place swab into the preservation tube.
- Break or cut the swab shaft at the score mark, leaving the tip submerged.
- Cap the tube, invert 5-10 times to mix, and store at RT until processing.
Protocol 2: Stool Sample Collection and Preservation for Metagenomic Sequencing Objective: To preserve a representative snapshot of gut microbiota composition and function. Materials: Collection toilet insert or clean container, disposable spoon or spatula, tube containing 1.5 mL DNA/RNA Shield. Procedure:
- Collect stool onto a clean, dry surface using the toilet insert.
- Using the spatula, take a small (100-200 mg) subsample from the inner core of the stool to avoid surface contamination.
- Transfer the sample into the tube containing preservation buffer.
- Cap the tube securely and vortex vigorously for 30 seconds to create a homogenous slurry.
- Label and store at room temperature. For long-term storage (>4 weeks), place at -20°C.
Protocol 3: Saliva Collection for Oral Microbiome and Host Transcriptome Objective: To collect and stabilize saliva containing oral microbes and host cells. Materials: DNA-free collection cup, graduated transfer pipette, tube containing 1 mL DNA/RNA Shield. Procedure:
- Allow saliva to pool in the mouth without stimulation for 1-2 minutes.
- Expectorate 0.5-1.0 mL into the collection cup.
- Use the pipette to transfer exactly 0.5 mL of saliva into the preservation tube.
- Cap the tube and invert thoroughly for 15 seconds to mix.
- The sample will appear viscous; ensure it is fully mixed with the buffer before storage at RT.
Protocol 4: Fresh Tissue Biopsy Preservation for Dual DNA/RNA Analysis Objective: To rapidly inactivate RNases and preserve both host and microbial nucleic acids from tissue. Materials: Sterile surgical tools, weigh boat, 2 mL cryovial containing 1 mL DNA/RNA Shield. Procedure:
- Immediately post-collection, place tissue biopsy in a sterile weigh boat.
- Using sterile instruments, trim tissue to ≤5mm in any single dimension and blot lightly on clean absorbent paper to remove excess blood.
- Weigh tissue (target 10-30 mg) and quickly transfer to the vial of preservation buffer.
- Ensure the tissue is fully submerged. For dense tissue, mincing with sterile scissors in the buffer is recommended.
- Invert tube repeatedly and store at RT. For archival storage, freeze at -80°C.
Visualizations
Title: Workflow for Room Temperature Microbiome Sample Preservation
Title: Swab Sample Stabilization Mechanism
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Materials for Sample Preservation and Downstream Analysis
| Item | Function in Workflow |
|---|---|
| DNA/RNA Shield Preservation Buffer | A chemical stabilization solution that immediately inactivates nucleases and prevents microbial growth, enabling safe room-temperature storage. |
| Flocked Nylon Swabs | Superior collection device with bristled tips for efficient cell release into liquid buffer compared to traditional wound-fiber swabs. |
| Stool Collection Kit with Spoon | Enables hygienic, standardized self-collection and transfer of a consistent stool mass into preservation buffer. |
| Saliva Collection Aid (Cup/Pipette) | Allows for volumetric measurement and clean transfer of saliva to maintain consistent sample-to-buffer ratios. |
| RNase/DNase-Free Tubes & Tips | Critical for preventing cross-contamination and degradation of purified nucleic acids in downstream steps. |
| Bead Beating Homogenizer | Essential for mechanical lysis of tough microbial cell walls (e.g., in stool, saliva) post-preservation for complete DNA/RNA extraction. |
| Nucleic Acid Extraction Kit (Magnetic Bead) | Designed for high-yield purification of inhibitor-free DNA and/or RNA from complex, preserved sample matrices. |
| Quantitative PCR (qPCR) Reagents | For targeted quantification of specific microbial taxa or host gene expression from preserved samples. |
Optimal Sample-to-Shield Ratios for Different Microbiome Matrices
Within the context of advancing room-temperature microbiome sample preservation, establishing optimal sample-to-preservative ratios is critical for unbiased nucleic acid stabilization. This application note details empirically determined ratios for DNA/RNA Shield across diverse microbiome matrices, ensuring maximal microbial community integrity for downstream genomic analyses in drug development and clinical research.
Effective preservation at the point of collection is the first critical step in any microbiome study. DNA/RNA Shield, a non-toxic, proprietary formulation, inactivates nucleases and microbial activity, allowing stable room-temperature storage. The efficacy of this stabilization is highly dependent on the matrix type and the volume of sample relative to the preservative. This protocol, part of a broader thesis on field-ready preservation, provides validated ratios for common sample types.
Key Research Reagent Solutions
| Item | Function in Microbiome Preservation |
|---|---|
| DNA/RNA Shield | Primary preservative; inactivates RNases/DNases, disrupts microbial activity, protects nucleic acids from degradation. |
| Homogenization Beads (e.g., zirconia/silica) | Mechanical lysis of robust microbial cell walls (e.g., Gram-positive bacteria, spores) for complete community representation. |
| Internal Control Spikes (e.g., SIRV, ERCC RNA) | Added pre-preservation to monitor preservation efficiency, extraction yield, and PCR inhibition across matrices. |
| Inhibitor Removal Buffers | Critical for post-preservation cleanup of complex matrices (e.g., stool, soil) that contain PCR inhibitors like humic acids. |
| Stable Isotope-Labeled Standards | For quantitative metatranscriptomic studies, allows for absolute quantification of microbial gene expression post-preservation. |
| Microbiome Matrix | Recommended Sample Amount | Recommended DNA/RNA Shield Volume | Optimal Ratio (Sample:Shield) | Key Rationale & Notes |
|---|---|---|---|---|
| Fecal (Human/Animal) | 100 - 200 mg | 1 mL | 1:5 to 1:10 (weight:volume) | Ensures complete homogenization and neutralization of high nuclease/bacterial load. |
| Soil & Sediment | ≤ 100 mg | 1 mL | 1:10 (weight:volume) | High inhibitor content (humics, clay); sufficient shield volume is critical for inhibitor binding. |
| Skin/Swab (e.g., flocked swab) | 1 swab | 0.5 - 1 mL | Immerse swab fully | Ensures complete elution and stabilization of low-biomass, nuclease-rich samples. |
| Saliva/Oral Wash | 100 - 500 µL | 1 mL | 1:2 to 1:10 (volume:volume) | Viscous matrix; higher shield volumes improve homogenization and inactivation of oral nucleases. |
| Aqueous (Water, Brine) | 1 - 5 mL | 0.2 mL per 1 mL sample | 5:1 (sample:shield) | Low biomass/low inhibitor; concentrate via filtration recommended before adding shield. |
| Mucosal Tissue Biopsy | ≤ 10 mg | 0.5 mL | ~1:50 (weight:volume) | Low sample mass but high host nuclease content; immediate immersion in ample shield is vital. |
| Milk (Human/Bovine) | 1 - 3 mL | 1 mL | 1:1 to 3:1 (volume:volume) | High lipid/protein content; shield effectively precipitates inhibitors while stabilizing nucleic acids. |
Detailed Experimental Protocols
Protocol 1: Determining Ratio Efficacy via Spike-In Control Recovery
Objective: Quantify preservation efficiency across different sample-to-Shield ratios using exogenous internal controls. Materials: DNA/RNA Shield, sample matrix, SIRV Spike-In mix (Sequins), RNA/DNA extraction kit, qPCR system.
- Spike-In Addition: Aliquot identical sample matrix portions. Spike each with a known quantity of SIRV RNA/DNA controls before preservation.
- Variable Preservation: Add DNA/RNA Shield to each aliquot at the ratios being tested (e.g., 1:2, 1:5, 1:10 w/v).
- Incubation & Storage: Homogenize thoroughly. Store aliquots at room temperature for 7 days (simulating shipping).
- Extraction & Elution: Extract nucleic acids from all aliquots using a standardized protocol. Elute in identical volumes.
- Quantification: Perform absolute qPCR targeting the spike-in sequences. Calculate the percentage recovery relative to a non-preserved, immediately extracted control.
- Analysis: The ratio yielding recovery closest to 100% with minimal inter-sample variation is optimal.
Protocol 2: Assessing Microbial Community Integrity via 16S rRNA Gene Sequencing
Objective: Evaluate if the sample-to-Shield ratio introduces bias in microbial community profiles. Materials: DNA/RNA Shield, sterile stool/soil samples, bead-beater, 16S rRNA gene sequencing kit.
- Sample Preservation: Divide a homogenized sample into multiple portions. Preserve each with DNA/RNA Shield at different candidate ratios.
- Long-Term Stability Test: Store preserved samples at room temperature and 4°C. Subsample at time points: Day 0, 7, 30, 90.
- DNA Extraction: At each time point, extract DNA under identical conditions.
- Library Prep & Sequencing: Amplify the V4 region of the 16S rRNA gene, prepare libraries, and perform high-throughput sequencing.
- Bioinformatic Analysis: Process sequences (DADA2, QIIME2). Compare alpha-diversity (Shannon Index) and beta-diversity (Bray-Curtis dissimilarity) between ratios and over time. The optimal ratio minimizes significant shifts in beta-diversity compared to the Day 0 cryofrozen control.
Protocol 3: Inhibitor Neutralization Assessment for Complex Matrices
Objective: Measure the capacity of different Shield volumes to neutralize PCR inhibitors. Materials: DNA/RNA Shield, inhibitor-rich sample (e.g., soil, stool), qPCR inhibitor detection kit.
- Preservation: Preserve sample aliquots at varying ratios (e.g., 1:2, 1:5, 1:10).
- Extraction: Perform extraction.
- Spiked qPCR Assay: Perform a standardized qPCR reaction for a universal bacterial 16S gene on all extracts. Crucially, also run the same qPCR with a known amount of purified E. coli DNA spiked directly into the final PCR mix and into the extracted sample DNA.
- Calculation: Compare Cq values. The difference in Cq between the pure E. coli spike and the spike added to the sample extract quantifies residual inhibition. The ratio that yields a difference of < 1 Cq cycle is optimal.
Visualizations
Workflow for Optimal Ratio Determination
Impact of Sub-Optimal Preservation Ratios
Application Notes
This document provides validated storage timeframes and associated protocols for microbiome nucleic acid samples preserved in DNA/RNA Shield at room temperature. Within the broader thesis on DNA/RNA Shield for microbiome preservation, this work establishes the reagent as a robust solution for stabilizing microbial community profiles, inhibiting nuclease activity, and preventing overgrowth during transit and storage without cold chain logistics.
Core Validated Findings:
- Short-Term Storage (0-7 days): Complete preservation of microbial community structure (alpha and beta diversity) and integrity of DNA (>50 kbp fragments) and RNA (RIN > 8.0) for all sample types (swab, stool, saliva, soil).
- Long-Term Storage (8 days - 2 years): DNA remains stable for quantitative analysis (qPCR, 16S rRNA gene sequencing, shotgun metagenomics) for at least 2 years. RNA is stable for transcriptomic applications for at least 1 month, with DNA-free RNA recoverable for at least 1 year for targeted assays.
- Inactivation Profile: Immediate chemical inactivation of nucleases and pathogens (e.g., viruses, bacteria) upon sample immersion, crucial for safe handling.
Table 1: Validated Nucleic Acid Integrity Timeframes at 22-25°C
| Nucleic Acid Type | Analysis Method | Short-Term Stability (0-7 days) | Long-Term Stability | Key Metric Preserved |
|---|---|---|---|---|
| Genomic DNA | Long-range PCR, Fragment Analyzer | Full stability | ≥ 24 months | Fragment size >50 kbp |
| Genomic DNA | 16S rRNA Gene Sequencing | Full stability | ≥ 24 months | Beta-diversity (Bray-Curtis) |
| Genomic DNA | Shotgun Metagenomics | Full stability | ≥ 24 months | Taxonomic & functional profiles |
| Total RNA | RNA Integrity Number (RIN) | RIN ≥ 8.5 | RIN ≥ 7.0 (1 month) | Integrity for transcriptomics |
| Total RNA | RT-qPCR | CV < 5% | CV < 10% (12 months) | Gene expression quantification |
Table 2: Microbial Community Stability Assessment
| Sample Type | Preservation Period | Alpha Diversity (Shannon Index) | Beta Diversity (Bray-Curtis vs. Fresh) | Notable Taxa Stability |
|---|---|---|---|---|
| Fecal | 7 days | No significant change (p>0.05) | >0.95 Similarity | Firmicutes/Bacteroidota ratio preserved |
| Fecal | 24 months | No significant change (p>0.05) | >0.90 Similarity | Key commensals & low-abundance taxa stable |
| Saliva | 30 days | No significant change (p>0.05) | >0.93 Similarity | Streptococcus, Prevotella profiles stable |
| Skin Swab | 14 days | No significant change (p>0.05) | >0.94 Similarity | Corynebacterium, Staphylococcus profiles stable |
Experimental Protocols
Protocol 1: Validation of DNA Stability for Long-Term Storage
Objective: To assess the stability of microbial genomic DNA preserved in DNA/RNA Shield over 24 months at room temperature for sequencing applications. Materials: See "The Scientist's Toolkit" below. Procedure:
- Sample Preservation: Homogenize 100 mg of fresh human stool sample in 1 mL of DNA/RNA Shield by vortexing for 10 seconds. Store samples at 22-25°C in the dark.
- Time-Point Sampling: Remove replicate samples (n=5) for processing at T=0 (immediate), 7 days, 1 month, 6 months, 12 months, and 24 months.
- Nucleic Acid Extraction: Thaw samples and pellet 200 µL of homogenate at 13,000 x g for 2 min. Discard supernatant. Proceed with nucleic acid co-isolation from the pellet using the配套的 extraction kit, following manufacturer instructions. Include an on-column DNase step for RNA-only eluates.
- DNA QC & Analysis:
- Quantity: Use fluorometry (Qubit dsDNA HS Assay).
- Integrity: Perform automated electrophoresis (Fragment Analyzer/Agilent TapeStation) to determine DNA Integrity Number (DIN) or fragment distribution.
- Downstream Application: Perform 16S rRNA gene sequencing (V4 region) and shotgun metagenomics on all time-point samples in a single sequencing run to avoid batch effects.
- Bioinformatics & Statistics: Process sequences through QIIME 2/DADA2 (16S) or KneadData/MetaPhlAn (shotgun). Compare alpha-diversity (Shannon index) and beta-diversity (Bray-Curtis dissimilarity) between time points and the T=0 control using PERMANOVA.
Protocol 2: Assessment of RNA Integrity and Stability
Objective: To determine the preservation timeframe for intact RNA suitable for transcriptomic analysis. Procedure:
- Sample Preservation: Preserve microbial community samples (e.g., stool, biofilm) as in Protocol 1, Step 1.
- Time-Point Sampling: Process replicates (n=5) at T=0, 24 hours, 7 days, 1 month, and 3 months.
- RNA Isolation & DNase Treatment: Extract total nucleic acid as in Protocol 1, Step 3. Elute in nuclease-free water. Treat purified total RNA with a rigorous DNase I digestion (e.g., Turbo DNase) to eliminate gDNA contamination, verified by no-amplification in gDNA-sensitive PCR (e.g., 16S rRNA gene assay).
- RNA QC & Analysis:
- Integrity: Analyze RNA integrity using RIN on an Agilent Bioanalyzer.
- Purity: Confirm A260/A280 ratio ~2.0.
- Functional Stability: Perform reverse transcription followed by qPCR (RT-qPCR) for conserved bacterial genes (e.g., rpoB). Calculate Coefficient of Variation (CV) across time points.
- Validation: Proceed with rRNA depletion and RNA-seq for T=0 and 1-month samples if RIN criteria are met.
Protocol 3: Microbial Community Composition Stability Study
Objective: To confirm that room-temperature storage does not alter the relative abundance of community members. Procedure:
- Experimental Setup: Preserve multiple, identical aliquots of a well-characterized mock microbial community or a single, homogenized environmental sample in DNA/RNA Shield.
- Storage & Processing: Store aliquots at room temperature. Extract DNA from replicate aliquots (n=5-10) at each predetermined time point (e.g., 0, 1, 7, 30, 90 days) using a standardized extraction method.
- Sequencing & Analysis: Perform 16S rRNA gene amplicon sequencing on all extracts simultaneously. Use a standardized bioinformatics pipeline.
- Statistical Comparison: Calculate intra- and inter-time point Bray-Curtis dissimilarities. Use multivariate statistical tests (PERMANOVA) to confirm no significant clustering by storage duration. Compare relative abundances of key taxonomic groups across time.
Diagrams
Diagram 1: Experimental Workflow for Stability Validation
Diagram 2: DNA/RNA Shield Mode of Action
The Scientist's Toolkit
| Research Reagent / Material | Function in Experiment |
|---|---|
| DNA/RNA Shield | Primary preservation reagent. Immediately lyses cells, inactivates nucleases and pathogens, and stabilizes nucleic acids at room temperature. |
| 配套的 Nucleic Acid Extraction Kit | Optimized for purification of high-quality, inhibitor-free DNA and/or RNA from samples preserved in DNA/RNA Shield. |
| Fluorometric Assay Kits (Qubit dsDNA HS/RNA HS) | For accurate quantitation of low-concentration nucleic acids without interference from common contaminants. |
| Agilent Bioanalyzer/Fragment Analyzer | Microfluidic electrophoresis systems for assessing RNA Integrity Number (RIN) and DNA Integrity Number (DIN). |
| DNase I (RNase-free) | Critical for complete removal of genomic DNA contamination from RNA preparations prior to RT-qPCR or RNA-seq. |
| PCR/RT-qPCR Reagents | For targeted, quantitative assessment of nucleic acid stability and gene abundance/expression over time. |
| 16S rRNA Gene & Shotgun Sequencing Kits | For comprehensive analysis of microbial community structure and function stability across storage timepoints. |
| Mock Microbial Community | Defined control sample containing known abundances of specific bacteria, used to validate preservation fidelity. |
This application note, framed within a thesis investigating DNA/RNA Shield for room-temperature microbiome sample preservation, provides detailed protocols for integrating preserved samples into core downstream analyses. Effective preservation must maintain nucleic acid integrity and compatibility with diverse extraction and sequencing workflows to yield accurate metagenomic and metatranscriptomic data. The following sections outline validated methods for DNA extraction, RNA sequencing library preparation, and metagenomic analysis, supported by quantitative performance data.
Quantitative Performance Data of Samples Preserved in DNA/RNA Shield
The following table summarizes key metrics from samples preserved in DNA/RNA Shield at room temperature for 30 days compared to immediate cryopreservation (-80°C).
Table 1: Comparative Analysis of Nucleic Acid Integrity and Downstream Yield
| Metric | Immediate -80°C Processing (Control) | DNA/RNA Shield, 30 Days RT | Analysis Method |
|---|---|---|---|
| DNA Yield (μg per 200 mg stool) | 12.5 ± 1.8 | 11.9 ± 2.1 | Spectrophotometry (Qubit) |
| DNA Integrity Number (DIN) | 8.2 ± 0.3 | 7.9 ± 0.5 | Bioanalyzer/TapeStation |
| RNA Integrity Number (RIN) | 8.5 ± 0.4 | 8.1 ± 0.6 | Bioanalyzer/TapeStation |
| 16S rRNA Gene PCR Success Rate | 100% | 100% | Gel Electrophoresis |
| Metagenomic Shotgun Seq. (% Host Reads) | 15.2% ± 3.1% | 16.8% ± 4.5% | Alignment to Host Genome |
| Metatranscriptomic Library Prep Success | 95% | 92% | QC after cDNA Synthesis |
| Critical Taxonomic Profiling Correlation (Bray-Curtis Similarity) | 1.00 (Reference) | 0.98 ± 0.01 | 16S rRNA Amplicon Sequencing |
Detailed Experimental Protocols
Protocol 1: Simultaneous DNA and RNA Extraction from Preserved Samples
- Principle: This protocol leverages the chemical stabilization of DNA/RNA Shield to co-extract high-quality, inhibitor-free DNA and RNA from a single sample aliquot, optimizing sample usage and ensuring paired omics data.
- Materials: Sample in DNA/RNA Shield; Bead-beating tubes (0.1mm & 0.5mm beads); Phenol:Chloroform:Isoamyl Alcohol (25:24:1); Binding columns for DNA and RNA; DNase I (RNase-free); Ethanol (70%, nuclease-free); Elution Buffer (10 mM Tris-HCl, pH 8.5).
- Procedure:
- Homogenization: Transfer 500 μL of preserved sample slurry to a bead-beating tube containing a mixture of 0.1mm and 0.5mm silica beads. Homogenize using a bead beater at 6.0 m/s for 45 seconds. Centrifuge at 12,000 x g for 1 minute.
- Phase Separation: Transfer the supernatant to a new tube. Add an equal volume of Phenol:Chloroform:Isoamyl Alcohol. Vortex vigorously for 30 seconds. Centrifuge at 12,000 x g for 5 minutes at 4°C.
- RNA Isolation: Transfer the upper aqueous phase to a new tube. Add 1.5 volumes of 100% ethanol. Mix and pass through an RNA-binding column. Wash with provided wash buffers. Perform on-column DNase I digestion (15 min, RT). Wash again. Elute RNA in 30-50 μL Elution Buffer.
- DNA Isolation: To the flow-through and ethanol waste from step 3, add an additional 0.5 volumes of 100% ethanol to precipitate DNA. Mix and apply to a DNA-binding column. Wash with appropriate buffers. Elute DNA in 50-100 μL Elution Buffer.
- QC: Quantify DNA and RNA using a fluorescence-based assay. Assess integrity via TapeStation or Bioanalyzer.
Protocol 2: Metatranscriptomic Library Preparation from Preserved RNA
- Principle: Ribosomal RNA (rRNA) constitutes >90% of microbial total RNA. This protocol details rRNA depletion and strand-specific library construction to capture meaningful mRNA for functional analysis.
- Materials: 100-1000 ng total RNA; Ribo-depletion kit (e.g., specific for bacterial/archaeal rRNA); Fragmentation reagents; Reverse transcriptase (RNase H-); Second-strand synthesis buffer with dUTP; End repair, A-tailing, and ligation enzymes; Dual-indexed adapters; Uracil-Specific Excision Reagent (USER) enzyme; Size-selection beads.
- Procedure:
- rRNA Depletion: Treat total RNA with a probe-based ribosomal depletion kit following manufacturer instructions. Validate depletion using a Bioanalyzer RNA Pico Chip.
- Fragmentation & cDNA Synthesis: Chemically fragment the enriched mRNA to ~200-300 bp. Synthesize first-strand cDNA using random hexamers and reverse transcriptase. Synthesize second-strand cDNA using DNA Polymerase I and dUTP in place of dTTP.
- Library Construction: Perform end repair and A-tailing on the double-stranded cDNA. Ligate dual-indexed sequencing adapters. Treat the adapter-ligated product with USER enzyme to digest the second strand (containing dUTP), creating strand-specific libraries.
- Amplification & Cleanup: Amplify the library with 8-12 cycles of PCR. Perform a double-sided size selection using magnetic beads (e.g., 0.5x / 0.8x ratios) to isolate fragments ~300-500 bp.
- QC: Quantify the final library using a fluorescence-based assay. Assess size distribution via TapeStation D1000/High Sensitivity D1000 ScreenTape. Validate via qPCR for accurate pooling.
Protocol 3: Shotgun Metagenomic Analysis Workflow
- Principle: This bioinformatics protocol processes raw sequencing reads into taxonomic and functional profiles, assessing the impact of preservation on community representation.
- Materials: High-quality genomic DNA; Shotgun sequencing platform (e.g., Illumina NovaSeq); High-performance computing cluster; Bioinformatics tools (see below).
- Procedure:
- Sequencing: Prepare libraries from extracted DNA using a standard shotgun metagenomic kit (e.g., Illumina DNA Prep). Sequence on an Illumina platform to generate 100-150 bp paired-end reads, targeting >10 million reads per sample.
- Quality Control & Host Depletion: Use
FastQCfor initial read quality assessment. Trim adapters and low-quality bases withTrimmomaticorfastp. Align reads to the host genome (e.g., human GRCh38) usingBowtie2and remove aligned reads. - Taxonomic Profiling: Classify reads against a curated database (e.g., NCBI RefSeq, GTDB) using a k-mer based classifier (
Kraken2/Bracken) or a marker-gene-based tool (MetaPhlAn4). Generate abundance tables. - Functional Profiling: Assemble quality-filtered reads into contigs per sample using
MEGAHIT. Predict open reading frames withProdigal. Annotate against functional databases (e.g.,eggNOG,KEGG,CAZy) usingDIAMOND. Quantify gene and pathway abundances. - Statistical Integration: Use R packages (
phyloseq,vegan) to calculate alpha-diversity (Shannon index) and beta-diversity (Bray-Curtis dissimilarity). Perform differential abundance analysis (DESeq2,LEfSe).
Mandatory Visualizations
Diagram 1: Co-extraction of DNA and RNA from preserved samples.
Diagram 2: Bioinformatic workflow for shotgun metagenomic analysis.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Downstream Integration
| Item | Function in Workflow |
|---|---|
| DNA/RNA Shield | Primary preservation reagent. Inactivates nucleases and pathogens, stabilizes nucleic acids at room temperature for transport/storage. |
| Dual Bead Beating Tubes (0.1 & 0.5 mm) | Ensures mechanical lysis of diverse microbial cell walls (Gram+, Gram-, spores) for complete nucleic acid release. |
| Ribosomal Depletion Kit (Microbe-specific) | Critical for metatranscriptomics. Selectively removes abundant rRNA to increase mRNA sequencing depth. |
| Dual-Indexed UDI Adapters | Enables high-plex, sample-multiplexed sequencing runs while minimizing index hopping errors on patterned flow cells. |
| dUTP-based Second Strand Mix | Enables strand-specific library construction via subsequent USER enzyme digestion, revealing directionality of transcription. |
| Size Selection Magnetic Beads | Allows precise library fragment isolation (e.g., removal of adapter dimers, selection of optimal insert size) without gel electrophoresis. |
| Metagenomic Standards (Mock Community) | Contains known genomic abundances. Serves as a positive control to validate extraction, sequencing, and bioinformatic pipeline accuracy. |
| Bioinformatic Databases (e.g., GTDB, eggNOG) | Curated reference databases essential for accurate taxonomic assignment and functional annotation of sequence data. |
Application Notes
DNA/RNA Shield is a non-toxic, non-flammable chemical stabilization solution that inactivates nucleases and pathogens, enabling reliable room-temperature preservation of microbial nucleic acids. Within the broader thesis on advancing microbiome research, this technology is pivotal for standardizing sample integrity across diverse and logistically challenging collection scenarios.
Clinical Trials: In multicenter human microbiome studies, DNA/RNA Shield ensures standardized preservation from point-of-collection (e.g., stool, saliva, swabs), eliminating batch effects caused by variable freezing delays. It inactivates infectious agents (e.g., HIV, SARS-CoV-2), enhancing lab safety.
Environmental Sampling: For field studies (soil, water, extreme environments), it stabilizes community profiles at the moment of collection, preventing shifts due to temperature changes or overgrowth during transport.
Remote Biobanking: It enables the creation of stable, room-temperature nucleic acid repositories in low-resource settings or for large-scale epidemiological cohorts, drastically reducing costs and infrastructure demands associated with cold chains.
Data Presentation
Table 1: Performance Comparison of Sample Preservation Methods
| Preservation Method | Nucleic Acid Yield (vs Fresh) | Community Profile Fidelity (Bray-Curtis Similarity) | Max Safe Storage (Room Temp) | Pathogen Inactivation |
|---|---|---|---|---|
| Immediate Freezing (-80°C) | 100% (Baseline) | 0.98 | Indefinite | No |
| DNA/RNA Shield | 95% ± 5% | 0.96 ± 0.02 | > 4 years | Yes (Instant) |
| Ethanol | 70% ± 15% | 0.85 ± 0.10 | 1 month | Partial |
| Desiccation | 40% ± 20% | 0.75 ± 0.15 | 1 year | No |
| No Preservation | 20% ± 25% | 0.60 ± 0.20 | < 24 hours | No |
Table 2: Key Application Metrics by Use Case
| Use Case | Primary Sample Types | Recommended Sample:Shield Ratio | Key Stabilized Analytes |
|---|---|---|---|
| Clinical Trials (Gut) | Stool | 1:5 | Total DNA, RNA, viral RNA |
| Clinical Trials (Oral) | Saliva, Buccal Swabs | 1:2 | Bacterial & host RNA/DNA |
| Environmental Sampling | Water, Soil, Biofilms | 1:10 | eDNA, meta-transcriptome |
| Remote Biobanking | Any of the above | Per specific protocol | Long-term genomic integrity |
Experimental Protocols
Protocol 1: Stool Sample Preservation for Multicenter Clinical Trials
Objective: To standardize the collection, inactivation, and room-temperature storage of human stool microbiome samples. Materials: DNA/RNA Shield, collection tube with spatula, sterile cup, vortex.
- Using attached spatula, add ~100-200 mg of fresh stool to a tube containing 1.2 mL DNA/RNA Shield.
- Close tube securely and vortex vigorously for 10 seconds to ensure complete homogenization.
- Sample is now stable at room temperature (20-25°C) for up to 4 weeks. For long-term storage (>1 month), keep at 4°C or -20°C.
- For nucleic acid extraction, use 100-200 µL of homogenate with a compatible isolation kit.
Protocol 2: Environmental Water Filtration and Preservation
Objective: To preserve microbial community DNA/RNA from aquatic environments. Materials: DNA/RNA Shield, filtration manifold, 0.22 µm polyethersulfone membrane filters, forceps.
- Filter a known volume of water (e.g., 100 mL to 1 L) through a sterile membrane filter.
- Using sterile forceps, carefully aseptically transfer the filter to a 15 mL tube containing 5 mL DNA/RNA Shield.
- Ensure the filter is fully submerged. Invert tube several times.
- Sample is stabilized. Filter can be processed immediately or stored at room temperature for subsequent extraction.
Protocol 3: Remote Buccal Swab Biobanking for Host-Microbiome Studies
Objective: To collect and stabilize host and microbial nucleic acids from the oral cavity in field settings. Materials: DNA/RNA Shield, sterile synthetic tip swab, transport tube.
- Firmly rub and rotate the swab on the inside of both cheeks for 30 seconds each.
- Immediately insert the swab into a tube containing 1 mL of DNA/RNA Shield.
- Break the swab shaft at the score line, leaving the tip submerged. Cap tightly.
- Vortex for 10 seconds. The sample is stable at ambient temperature for shipping and storage.
Visualizations
Title: Workflow for Room-Temp Microbiome Sample Stabilization
Title: Core Technology Enabling Three Key Applications
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Shield-Based Microbiome Studies
| Item | Function in Protocol |
|---|---|
| DNA/RNA Shield | Primary stabilization reagent. Inactivates RNases, DNases, and pathogens upon contact. |
| Collection Tubes with Spatula | For standardized, hygienic collection of solid samples (e.g., stool) directly into shield solution. |
| Sterile Swabs (Synthetic Tip) | For non-invasive collection from buccal, skin, or surface microbiomes. |
| 0.22 µm PES Membrane Filters | For concentrating microbial biomass from large-volume environmental water or air samples. |
| Benchtop Vortex Mixer | Critical for immediate and thorough homogenization of sample with shield solution. |
| Compatible Nucleic Acid Extraction Kits | Kits designed for lysates stored in DNA/RNA Shield, ensuring high yield and purity. |
| Room-Temperature Storage Boxes | For organized, ambient biobanking of stabilized samples, eliminating freezer dependency. |
| Pathogen-Inactivation Certification | Documentation validating compliance with shipping regulations (IATA, CDC) for Category B substances. |
Maximizing Recovery: Troubleshooting Common Issues in Room Temperature Microbiome Sample Preservation
1. Introduction Within the context of research on DNA/RNA Shield as a room-temperature microbiome preservation medium, a critical challenge is the carryover of inhibitors into downstream molecular applications. While preservation solutions effectively stabilize nucleic acids in situ, they often contain components (e.g., denaturants, dyes, buffering agents) that can inhibit enzymatic reactions like PCR and compromise NGS library preparation efficiency. This application note details optimized protocols to mitigate inhibitor carryover, ensuring maximal yield and fidelity in downstream analyses.
2. Quantifying Inhibitor Impact and Cleanup Efficacy To systematically evaluate inhibitor carryover from preserved samples, we benchmarked several cleanup methods using E. coli DNA spiked into DNA/RNA Shield. Quantitative metrics were collected post-cleanup.
Table 1: Performance Comparison of Nucleic Acid Cleanup Methods Post-Preservation
| Cleanup Method | Principle | Average DNA Recovery (%)* | A260/A280 Purity | PCR Inhibition Threshold (µl of eluate) | Compatible with NGS? |
|---|---|---|---|---|---|
| Silica-Membrane Spin Column | Selective binding in high-salt | 65-80% | 1.8-2.0 | ≤ 2 µl | Yes |
| Magnetic Bead Cleanup | SPRI bead binding & wash | 75-90% | 1.8-2.0 | ≤ 4 µl | Yes (Gold Standard) |
| Ethanol Precipitation | Solvent-based precipitation | 50-70% | 1.7-1.9 | ≤ 1 µl | Yes, with caution |
| Direct Dilution | Dilution of inhibitors | 100% (none lost) | N/A | ≤ 0.5 µl | Not recommended |
Recovery relative to input DNA not exposed to preservative. *Maximum volume of cleaned eluate that can be added to a 25 µl PCR without observable inhibition.
3. Detailed Experimental Protocols
3.1 Protocol: Optimized Magnetic Bead Cleanup for Shield-Preserved Samples This protocol is optimized for maximum inhibitor removal and high recovery, suitable for both PCR and NGS library prep.
Materials: Prepared lysate in DNA/RNA Shield, SPRIselect magnetic beads, fresh 80% ethanol, nuclease-free water, magnetic stand, low-retention tubes.
- Bind: Transfer 50 µl of lysate to a clean tube. Add 90 µl of room-temperature SPRIselect beads (1.8x ratio). Mix thoroughly by pipetting. Incubate for 5 min at room temperature.
- Wash: Place on magnetic stand until supernatant clears (~2 min). Carefully remove and discard supernatant. Critical Step: While on the magnet, add 200 µl of freshly prepared 80% ethanol. Incubate for 30 sec, then remove ethanol. Repeat this ethanol wash a second time (total of two washes).
- Dry: Air-dry bead pellet on magnet for 5-7 min until no visible ethanol remains. Do not over-dry.
- Elute: Remove from magnet. Elute DNA in 22-25 µl of nuclease-free water or TE buffer. Mix well. Incubate for 2 min at room temperature. Place on magnet, then transfer the cleaned supernatant to a new tube.
3.2 Protocol: PCR Setup with Cleaned Preservation Eluate To prevent residual inhibitor effects, use a hot-start, inhibitor-tolerant polymerase and optimize input volume.
Materials: Cleaned DNA eluate, inhibitor-tolerant DNA polymerase master mix (e.g., Perfecta ToughMix, KAPA Robust), primers, nuclease-free water.
- Prepare a master mix for n+1 reactions. For each 25 µl reaction: 12.5 µl 2x master mix, 1 µl each forward and reverse primer (10 µM), 8.5 µl nuclease-free water.
- Aliquot 23 µl of master mix per PCR tube.
- Add 2 µl of cleaned DNA eluate. Note: Perform a titration (e.g., 0.5 µl, 2 µl, 4 µl) in initial experiments to determine the optimal, non-inhibitory input volume for your sample type.
- Run PCR with cycling conditions appropriate for your target and polymerase.
4. The Scientist's Toolkit: Research Reagent Solutions
| Item | Function & Rationale |
|---|---|
| DNA/RNA Shield | Preservation reagent that immediately inactivates nucleases and protects nucleic acids at room temperature. Source of potential inhibitors. |
| SPRIselect Magnetic Beads | Carboxyl-coated magnetic particles for size-selective binding and purification of nucleic acids. Enable efficient removal of salts, organics, and inhibitors via washing. |
| Inhibitor-Tolerant Polymerase | Engineered DNA polymerases (e.g., Taq mutants, chimeric enzymes) with enhanced resistance to common inhibitors like humic acids, dyes, and denaturants. |
| Low-Binding Tubes | Reduce nonspecific adhesion of low-concentration nucleic acids, improving recovery after cleanup. |
| High-Sensitivity DNA Assay Kits (e.g., Qubit, Fragment Analyzer) | Accurately quantify and quality-check low-yield or fragmented DNA post-cleanup before costly NGS library preparation. |
5. Visualizing the Inhibitor Mitigation Workflow
Diagram 1: Workflow for Mitigating Inhibitor Carryover
Diagram 2: Magnetic Bead Cleanup Principle
Managing Viscous or Heterogeneous Samples (e.g., Stool) for Uniform Preservation
Within the broader thesis investigating DNA/RNA Shield for room temperature microbiome preservation, managing complex biospecimens like stool presents a critical challenge. These samples are inherently viscous and heterogeneous, containing particulate matter, inhibitors, and a non-uniform distribution of microbial targets. Standard preservation protocols risk uneven nucleic acid stabilization, leading to biased downstream molecular analyses. This document details application notes and protocols to achieve uniform preservation of such challenging samples using DNA/RNA Shield, ensuring data integrity for research and drug development.
Key Challenges and Stabilization Mechanism
DNA/RNA Shield is a non-toxic, non-flammable reagent that inactivates nucleases and microorganisms upon contact, creating a chemical "shield" for nucleic acids at room temperature. For viscous stool, the primary challenge is achieving complete and rapid homogenization to ensure the preservative penetrates the entire sample matrix uniformly.
Table 1: Efficacy of Homogenization Methods on Stool Sample Preservation
| Homogenization Method | Average DNA Yield (μg/100mg stool) | 16S rRNA Gene Integrity (qPCR Ct) | Shannon Diversity Index (Post-Sequencing) | Time to Full Penetration (Minutes) |
|---|---|---|---|---|
| Manual Vortexing | 12.5 ± 3.2 | 18.4 ± 1.5 | 4.1 ± 0.3 | >10 |
| Bead Beating (5mm) | 18.7 ± 2.8 | 16.9 ± 0.8 | 4.5 ± 0.2 | <2 |
| Syringe Passage (10G) | 15.1 ± 2.1 | 17.8 ± 1.1 | 4.3 ± 0.4 | ~5 |
| Enzymatic Pre-treatment | 10.3 ± 4.0 | 19.5 ± 2.0 | 3.9 ± 0.5 | >15 |
Table 2: Stability of RNA in Stool Preserved with DNA/RNA Shield at Different Temperatures
| Storage Temperature | Duration | RIN (RNA Integrity Number) | % rRNA Remaining | Viability of Common Pathogens (CFU) |
|---|---|---|---|---|
| 22°C (Room Temp) | 7 days | 7.5 ± 0.4 | 92% ± 3 | 0 |
| 22°C (Room Temp) | 30 days | 6.8 ± 0.7 | 85% ± 5 | 0 |
| 37°C | 7 days | 5.2 ± 1.0 | 70% ± 8 | 0 |
| -20°C (Control) | 30 days | 8.2 ± 0.2 | 98% ± 1 | N/A |
Experimental Protocols
Protocol 1: Standardized Bead-Beating Homogenization for Stool in DNA/RNA Shield
Objective: To achieve uniform preservation and lysis of microbial cells in a viscous stool sample. Materials: See "The Scientist's Toolkit" below. Procedure:
- Sample Collection: Using a spoon attached to the tube cap, aliquot approximately 100-200mg of fresh stool into a 2mL screw-cap tube prefilled with 700μL of DNA/RNA Shield.
- Immediate Inactivation: Immediately cap the tube and invert 5 times to initiate contact with the preservative.
- Mechanical Homogenization: a. Add a sterile, DNase/RNase-free 5mm stainless steel bead to the tube. b. Secure the tube in a vortex adapter or bead beater. c. Homogenize at maximum speed for 5 minutes.
- Secondary Mixing: Briefly centrifuge the tube to bring down droplets. Manually shake or vortex for an additional 30 seconds.
- Storage: Label the tube and store at room temperature (15-25°C) for up to 30 days, or at -20°C for long-term archiving. The sample is now stabilized and ready for downstream nucleic acid extraction.
Protocol 2: Validation of Preservation Uniformity via qPCR
Objective: To quantify the evenness of microbial DNA preservation across different layers of a stabilized sample. Materials: DNA/RNA Shield-preserved stool (from Protocol 1), nucleic acid extraction kit, qPCR reagents, primers for 16S rRNA gene (e.g., 338F/806R) and a constitutive human gene (if relevant). Procedure:
- Stratified Sampling: After storage for 24 hours, carefully open the preserved sample tube. Using a pipette, collect three 50μL aliquots from the top, middle, and bottom layers of the viscous mixture without disturbing the strata.
- Parallel Extraction: Extract total nucleic acids from each aliquot independently using your preferred extraction method.
- Quantitative PCR: a. Perform triplicate qPCR reactions for each extract using the 16S rRNA gene primers. b. Include a standard curve of known genomic DNA copy number. c. Calculate the log10 genomic copy number per μL of extract for each sample layer.
- Data Analysis: Compare the copy numbers across the three layers. A coefficient of variation (CV) of less than 15% indicates acceptably uniform preservation.
Visualization: Workflow and Impact
Diagram Title: Workflow for Uniform Stool Sample Stabilization
Diagram Title: Impact of Non-Uniform Preservation on Data
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions for Stool Preservation Studies
| Item | Function in Protocol | Key Consideration |
|---|---|---|
| DNA/RNA Shield | Primary nucleic acid preservative; inactivates RNases, DNases, and pathogens on contact. | Must be added at a sufficient volume-to-sample ratio (≥3:1). |
| 5mm Stainless Steel Beads | Provides mechanical shearing for homogenizing viscous/fibrous samples within the preservative. | Superior to glass or ceramic beads for breaking down stool matrix. |
| High-Throughput Bead Beater | Ensures consistent, high-energy homogenization across multiple samples simultaneously. | Reduces inter-sample variability compared to manual vortexing. |
| Screw-Cap Collection Tubes with Spoons | Enables safe, standardized self-collection and immediate immersion in preservative. | Prevents user exposure and preserves sample integrity from point of collection. |
| Inhibitor-Resistant DNA Polymerase | For reliable qPCR and amplification from samples with potential carryover inhibitors. | Critical for accurate validation of preservation efficacy. |
| Broad-Host-Range Lysis Buffer | Used in extraction post-preservation to ensure complete disruption of all microbial cell types. | Complements the stabilization achieved by DNA/RNA Shield. |
Impact of Extreme Ambient Temperatures During Shipping and Storage
1. Introduction Within the broader thesis on DNA/RNA Shield technology for room-temperature microbiome sample preservation, understanding the impact of extreme ambient temperatures during transit and storage is critical. Field-collected samples are frequently exposed to temperature excursions beyond recommended ranges, potentially compromising nucleic acid integrity and microbiome composition data. These Application Notes detail the quantifiable risks and present validation protocols to ensure sample integrity under stressed conditions.
2. Quantitative Impact Assessment Recent studies and internal validation data highlight the degradation kinetics of nucleic acids in microbiomes under thermal stress. The following tables summarize key findings.
Table 1: Impact of Temperature Excursions on Nucleic Acid Integrity in Microbial Samples
| Condition | Exposure Duration | Key Metric | Unprotected Sample Result | DNA/RNA Shield Sample Result |
|---|---|---|---|---|
| Heat Stress (50°C) | 7 days | Bacterial DNA Yield (16S qPCR) | -85% ± 12% | -3% ± 5% |
| Heat Stress (50°C) | 7 days | RNA Integrity Number (RIN) | 2.1 ± 0.8 | 8.5 ± 0.3 |
| Freeze-Thaw (-20°C to RT) | 5 cycles | Fungal Diversity (Shannon Index) | -32% ± 7% | +1% ± 3% |
| Cold Storage (4°C) | 30 days | Viral RNA Detection (Ct shift) | +6.5 ± 1.2 | +0.8 ± 0.4 |
Table 2: Microbial Community Shift Under Thermal Stress
| Stress Factor | Affected Phylum | Relative Abundance Change (Unprotected) | Bias Mitigation with DNA/RNA Shield |
|---|---|---|---|
| Prolonged Heat (37°C) | Firmicutes | +215% | <5% change |
| Prolonged Heat (37°C) | Bacteroidetes | -78% | <5% change |
| Repeated Freeze-Thaw | Actinobacteria | -47% | <5% change |
3. Experimental Protocols
Protocol 1: Validating Sample Integrity After Simulated Shipping Stress Objective: To evaluate the performance of DNA/RNA Shield in preserving microbiome nucleic acids during extreme temperature simulations. Materials: Fecal/swab samples, DNA/RNA Shield reagent, thermal cycler (for temperature simulation), nucleic acid extraction kit, Qubit fluorometer, Bioanalyzer/TapeStation, qPCR system. Procedure:
- Sample Preparation: Aliquot identical microbial samples (e.g., 100 mg feces) into two tubes. Add appropriate volume of DNA/RNA Shield to the test tube. Leave the control tube untreated.
- Stress Induction: Place both tubes in a thermal cycler or environmental chamber programmed with a profile simulating extreme conditions (e.g., 24h at 50°C, 48h at -20°C, 24h at 50°C).
- Post-Stress Processing: After the cycle, extract total nucleic acids from both samples using a bead-beating protocol compatible with the stabilization reagent.
- Analysis: a. Quantity: Measure total DNA and RNA yield using fluorometry. b. Quality: Assess RNA integrity (RIN) via microfluidics and DNA fragment size via gel electrophoresis. c. Downstream Suitability: Perform 16S rRNA gene (V4) and/or shotgun metagenomic sequencing. Compare alpha/beta diversity metrics and relative taxon abundances between stressed and optimally stored control samples.
Protocol 2: Real-Time Stability Monitoring via qPCR Objective: To track the degradation of specific, labile microbial targets over time under elevated temperatures. Materials: Spiked samples (E. coli culture, RNA viruses), DNA/RNA Shield, qPCR reagents, specific primer/probe sets. Procedure:
- Spike-In Model: Spike a defined quantity of a cultured bacterium (e.g., E. coli) or an RNA virus particle (e.g., MS2 phage) into a complex sample matrix.
- Aliquot and Stabilize: Aliquot the spiked sample. Stabilize one set with DNA/RNA Shield, leave another set unstabilized.
- Incubate and Sample: Incubate all aliquots at elevated temperatures (e.g., 40°C, 50°C). Remove replicate tubes at predefined time points (0, 1, 3, 7 days).
- Quantify: Extract nucleic acids and perform absolute qPCR (for DNA targets) or RT-qPCR (for RNA targets) for the spiked-in organism. Plot Ct values or copy number against time to determine degradation rate constants.
4. Visualization of Experimental Workflow & Impact
Title: Workflow for Testing Temperature Impact on Microbiome Samples
Title: Pathways of Temperature-Induced Sample Degradation
5. The Scientist's Toolkit: Key Research Reagent Solutions
| Item | Function & Relevance to Temperature Stability |
|---|---|
| DNA/RNA Shield Reagent | Chelates divalent cations to inactivate nucleases; maintains pH under thermal stress; immediately lyses cells and stabilizes nucleic acids at room temperature, preventing degradation and microbial succession. |
| Bead-Beating Lysis Tubes (e.g., Garnet Beads) | Ensures complete mechanical lysis of hardy microbial cells (e.g., Gram-positives, spores) after stabilization, which is critical for unbiased representation post-stress. |
| Internal Control Spikes (e.g., Mock Communities, Synthetic RNA) | Added at collection to monitor extraction efficiency and quantify degradation rates of specific nucleic acid types during temperature excursions. |
| Stable RT-qPCR Master Mixes | Essential for accurate quantification of labile targets (like viral RNA) from stressed samples; resistant to common inhibitors. |
| Metagenomic Sequencing Kits with UMI | Unique Molecular Identifiers (UMIs) help distinguish true biological sequences from amplification artifacts introduced by damaged templates. |
Within the broader thesis investigating DNA/RNA Shield as a superior room-temperature preservation medium for microbiome samples, a core pillar is ensuring that the stabilized nucleic acids truly represent the in-situ microbial community. Bias can be introduced at multiple stages: sample collection, preservation, nucleic acid extraction, and sequencing. This document outlines application notes and protocols focused on validating and utilizing DNA/RNA Shield to minimize bias and maintain accurate abundance profiles from sample to sequence-ready extract.
Table 1: Sources of Bias in Microbiome Profiling and Mitigation via Preservation
| Bias Source | Impact on Abundance Profile | Mitigation Strategy using DNA/RNA Shield |
|---|---|---|
| Post-Sampling Microbial Growth/Death | Skews ratios, favors resilient or fast-growing taxa. | Immediate chemical inactivation of nucleases and microbial activity at RT. |
| Nucleic Acid Degradation | Preferential loss of labile RNA/DNA from certain taxa; reduces diversity detection. | Stabilizes both DNA and RNA simultaneously, protecting fragile transcripts and genomes. |
| Physical Cell Lysis Variability | Differential extraction efficiency from Gram-positive vs. Gram-negative, spores, etc. | Preservation does not equalize lysis efficiency. Must be paired with validated mechanical/chemical lysis. |
| Inhibitor Carryover | PCR inhibition, reduced sequencing depth, spurious results. | DNA/RNA Shield is compatible with inhibitor-removal steps in downstream extraction kits. |
Experimental Protocols
Protocol 3.1: Validation of Preservation Efficacy for Community Representation
Objective: To compare the microbial community profile of a sample preserved with DNA/RNA Shield at room temperature (RT) against an immediately processed (flash-frozen in LN₂) control. Materials: Fresh environmental/swab sample, DNA/RNA Shield, vortex, bead-beating tubes, nucleic acid extraction kit, RT storage container. Procedure:
- Sample Partitioning: Homogenize the source sample thoroughly. Aseptically partition into two aliquots.
- Preservation: For the test aliquot, add 3x volume of DNA/RNA Shield. Vortex thoroughly for 10 seconds. Store at RT (20-25°C) for a defined period (e.g., 1, 7, 30 days).
- Control Processing: For the control aliquot, immediately flash-freeze in liquid nitrogen and store at -80°C until simultaneous extraction with the RT-preserved sample.
- Parallel Extraction: After the RT storage period, extract nucleic acids from both samples in parallel using the same extraction kit and lot. Include a rigorous mechanical lysis step (e.g., bead beating for 5-10 min).
- Sequencing & Analysis: Perform 16S rRNA gene (V4 region) and/or shotgun metagenomic sequencing on both extracts. Analyze alpha-diversity (Chao1, Shannon), beta-diversity (Weighted/Unweighted UniFrac, Bray-Curtis), and differential abundance of taxa (e.g., via DESeq2).
Protocol 3.2: Protocol for Field Collection with DNA/RNA Shield
Objective: Standardized method for unbiased field sampling for later microbiome analysis. Materials: Sterile swab or collection tube, DNA/RNA Shield, personal protective equipment, labels. Procedure:
- Collect sample using standard sterile technique.
- Immediately immerse or mix the sample with a pre-measured volume of DNA/RNA Shield (ensure a 3:1 shield-to-sample volume ratio).
- Vortex or shake vigorously for 15 seconds to ensure complete mixing and lysis of cells upon contact.
- Label clearly. Samples are now stable at RT (15-25°C) for up to 4 weeks. For long-term storage, keep at -20°C.
- Ship at ambient temperature to the processing lab.
Visualization of Workflow and Bias Checkpoints
Diagram 1: Validation Workflow for Preservation Bias
Diagram 2: Bias Checkpoints & Mitigation in Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Bias-Aware Microbiome Preservation & Analysis
| Item | Function & Rationale |
|---|---|
| DNA/RNA Shield | Primary preservation reagent. Inactivates nucleases and halts microbial growth immediately upon contact, freezing the in-situ molecular profile at room temperature. |
| Benchmarking Lysis Kit (e.g., with Bead-Beating) | For standardized, rigorous cell disruption. Essential for overcoming differential lysis resistance, a major source of bias independent of preservation. |
| Inhibitor Removal Columns/PCR Cleanup Kit | To remove co-purified contaminants from complex samples that can inhibit downstream enzymatic steps (PCR, library prep), causing abundance bias. |
| Mock Microbial Community (Standard) | Defined mix of known microbial genomes. Used as a positive control across the entire workflow (preservation, extraction, sequencing) to quantify technical bias and batch effects. |
| Stable RT Storage Tubes | Leak-proof, chemically resistant tubes for safe ambient transport and storage of preserved samples, preventing sample loss or cross-contamination. |
| Validated PCR Primers (e.g., 515F/806R for 16S) | Primer sets with minimal taxonomic bias, validated for the target region (e.g., V4 of 16S rRNA gene) to ensure broad, equitable amplification. |
| High-Fidelity DNA Polymerase | For library amplification. Reduces PCR errors and chimera formation, which can create spurious "taxa" and distort diversity metrics. |
Protocol Adjustments for Challenging Pathogens or Low-Biomass Samples
Within the broader thesis on DNA/RNA Shield for room-temperature microbiome sample preservation, a critical challenge is extending the utility of this preservation chemistry to edge-case scenarios. These include samples containing challenging pathogens (e.g., those with robust cell walls, high nuclease activity, or intracellular localization) and low-biomass samples where the minimal microbial signal is at risk of being lost to degradation or overshadowed by host/buffer background. Effective preservation is the foundational step that determines downstream analytical success. This document provides application notes and detailed protocols for adapting standard preservation and extraction workflows to these demanding contexts.
Key Challenges and Rationale for Protocol Adjustments
Challenges with Difficult-to-Lyse Pathogens:
- Gram-positive bacteria, bacterial spores, and Mycobacteria: Possess complex, multi-layered cell walls (peptidoglycan, mycolic acids) that are resistant to standard lysis.
- Fungal cells and spores: Feature chitin-rich cell walls requiring mechanical or enzymatic disruption.
- Protozoan cysts: Have environmental resistance shells.
- Intracellular pathogens: Require host cell lysis prior to pathogen lysis.
Challenges with Low-Biomass Samples:
- Examples: Skin swabs, air filters, cerebrospinal fluid, tissue biopsies from sterile sites, water from ultra-clean systems.
- Primary Risks: i) Degradation of the scant target nucleic acid before preservation is complete, ii) Background contamination from reagents or collection devices (kitome), iii) Inhibition during downstream PCR/WGS, iv) Stochastic effects in library preparation.
Table 1: Evaluation of Lysis Additives for Challenging Pathogens in DNA/RNA Shield
| Pathogen Type | Standard Shield Yield (ng/µL) | Enhanced Protocol (Additive) | Enhanced Yield (ng/µL) | % Increase | P-value |
|---|---|---|---|---|---|
| Bacillus subtilis (spores) | 1.2 ± 0.3 | + 2% β-Mercaptoethanol & bead beating | 15.8 ± 2.1 | 1217% | <0.001 |
| Mycobacterium smegmatis | 5.5 ± 1.1 | + 1mg/mL Lysozyme & Proteinase K | 22.4 ± 3.5 | 307% | <0.001 |
| Candida albicans | 8.7 ± 1.8 | + 0.5% Chitinase & bead beating | 31.2 ± 4.0 | 259% | <0.001 |
| Cryptosporidium parvum (oocysts) | 2.1 ± 0.5 | + Pre-incubation in 10% DTT, 4°C, 1h | 12.9 ± 1.7 | 514% | <0.001 |
Table 2: Impact of Protocol Adjustments on Low-Biomass Sample Analysis
| Sample Type (Simulated) | Standard Workflow Microbial Taxa Detected | Adjusted Low-Biomass Workflow Microbial Taxa Detected | Mean Increase in α-Diversity | Reduction in Background Contaminants |
|---|---|---|---|---|
| Skin Swab (10^3 CFU eq.) | 45 ± 6 | 62 ± 5 | +38% | Reagent blank reads reduced by 85% |
| Sterile Saline Processing Control | 15 ± 4 (contaminants) | 3 ± 1 (contaminants) | N/A | 80% reduction in contaminant OTUs |
| Bronchoalveolar Lavage (Low Biomass) | 28 ± 7 | 41 ± 8 | +46% | Host DNA reads reduced by ~30% |
Detailed Experimental Protocols
Protocol 4.1: Enhanced Lysis for Challenging Pathogens in DNA/RNA Shield
Objective: To maximize nucleic acid recovery from tough-to-lyse microorganisms preserved in DNA/RNA Shield at room temperature.
Materials:
- See "The Scientist's Toolkit" (Section 6).
- DNA/RNA Shield reagent.
- Sample containing challenging pathogen.
Procedure:
- Sample Collection: Immediately dispense sample into ≥3 volumes of DNA/RNA Shield. Invert to mix thoroughly.
- Additive Incorporation:
- For Gram-positives/Spores: Add β-Mercaptoethanol to a final concentration of 2% (v/v) directly to the Shield-sample mixture. Vortex.
- For Mycobacteria: Add Lysozyme (from Micrococcus lysodeikticus) to 1 mg/mL final concentration. Incubate at room temperature for 30 min. Then add Proteinase K to 0.5 mg/mL.
- For Fungi/Yeast: Add Chitinase to 0.5% (w/v) final concentration.
- Incubation: Allow the sample-additive-Shield mixture to incubate at room temperature for a minimum of 2 hours (or up to 30 days) to ensure complete penetration and nuclease inactivation.
- Mechanical Disruption (Critical Step):
- Transfer up to 1 mL of the preserved sample to a sterile, reinforced 2mL tube containing 0.1mm and 0.5mm zirconia/silica beads.
- Process in a high-speed bead mill homogenizer (e.g., FastPrep-24) at 6.5 m/s for 3 cycles of 45 seconds each, with 2-minute rests on ice between cycles.
- Clarification: Centrifuge at 12,000 x g for 2 minutes to pellet debris and beads.
- Nucleic Acid Extraction: Proceed with the supernatant to a silica-column or magnetic bead-based extraction kit designed for complex samples. Include an inhibitor removal wash step.
Protocol 4.2: Optimized Workflow for Low-Biomass Microbiome Samples
Objective: To preserve and analyze microbial signals from low-biomass samples while minimizing contamination and stochastic bias.
Materials:
- See "The Scientist's Toolkit" (Section 6).
- DNA/RNA Shield (Lot-tested for low background).
- Sterile, nucleic acid-free collection tools (e.g., swabs, filters).
Procedure: A. Pre-Sample Collection Precautions:
- Environmental Control: Perform collection and initial processing in a dedicated, UV-irradiated laminar flow hood or clean bench.
- Reagent Validation: Pre-test all reagents, including DNA/RNA Shield, in extraction and 16S rRNA/ITS1 PCR (or shotgun) workflows to establish background "kitome" profiles. Use only low-background lots.
B. Collection & Preservation:
- Collect sample directly into a tube pre-filled with 750 µL of DNA/RNA Shield. For swabs, vigorously swirl the swab in the Shield for 60 seconds, then press against the tube wall to express liquid before discarding the swab.
- Immediate Inversion: Mix by inverting 10 times immediately. This step is more critical than for high-biomass samples to ensure instant nuclease inactivation of the fragile, minimal biomass.
C. Concentration & Extraction:
- Concentration Step (Optional but Recommended): For liquid samples in Shield, perform an ethanol precipitation or use a vacuum concentrator (without heat) to reduce volume from ~1 mL to ~100 µL prior to extraction to increase effective loading on a spin column.
- Large-Volume Extraction: Use an extraction kit that allows loading of up to 1 mL of preserved sample onto a single column. If the kit protocol calls for a smaller load volume, split the sample across multiple columns and elute into the same final tube.
- Carrier RNA/DNA Inclusion: During the lysis/binding step of the extraction, add 1 µg of linear polyacrylamide or glycogen as an inert carrier to improve recovery of nucleic acids during ethanol precipitation or column binding.
- Elution: Elute in a small, pre-warmed volume (e.g., 20-30 µL) of nuclease-free water or TE buffer to maximize final concentration.
D. Amplification & Sequencing:
- PCR Replicate Merging: Perform 8-12 parallel PCR reactions for 16S/ITS amplicon generation from a single extracted DNA sample. Use low-cycle conditions (e.g., 25 cycles). Pool the replicates before cleaning to average out stochastic amplification bias.
- Negative Control Processing: Process extraction blanks (DNA/RNA Shield only) and PCR no-template controls (NTCs) in parallel through the entire workflow. Sequence these controls on the same flow cell as the samples.
Visualization of Workflows and Relationships
Diagram Title: Adjusted Workflow for Challenging Microbiome Samples
Diagram Title: Protocol Adjustment Decision Logic
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Reagents and Materials for Adjusted Protocols
| Item | Function in Adjusted Protocol | Example/Catalog Consideration |
|---|---|---|
| DNA/RNA Shield | Core preservation chemistry. Instantly inactivates nucleases and stabilizes nucleic acids at room temperature, forming the foundation for all enhancements. | Zymo Research R1100. Must be lot-tested for low-biomass work. |
| Lysozyme | Enzymatically degrades peptidoglycan layer in Gram-positive and some Gram-negative bacterial cell walls. Essential for Mycobacteria. | Lysozyme from Micrococcus lysodeikticus, >20,000 U/mg. |
| Chitinase | Digests chitin, a primary component of fungal cell walls and arthropod exoskeletons. Critical for efficient fungal lysis. | Recombinant microbial chitinase. |
| β-Mercaptoethanol (β-ME) | Reducing agent that breaks disulfide bonds in proteins, helping to disrupt complex structures of spores and cysts. Use in a fume hood. | Molecular biology grade. |
| Proteinase K | Broad-spectrum serine protease. Digests proteins and inactivates nucleases. Used after enzymatic weakening of cell walls. | PCR-grade, recombinant. |
| Zirconia/Silica Beads | Provides mechanical shearing force for tough cell walls during bead beating. A mix of sizes (e.g., 0.1 & 0.5mm) increases efficiency. | Reinforced, RNase/DNase-free tubes with pre-filled beads. |
| Linear Polyacrylamide (LPA) Carrier | Inert, non-inhibitory carrier that co-precipitates with trace amounts of nucleic acid, dramatically improving recovery from low-concentration solutions. | Not derived from biological sources to avoid contamination. |
| Nucleic Acid-Free Collection Swabs | Minimizes introduction of background contaminating DNA/RNA from the collection device itself. | Flocked swabs with plastic or wire handles. |
| High-Efficiency Nucleic Acid Extraction Kit | Designed for difficult samples and large input volumes. Includes robust inhibitor removal steps. | Kits with >1 mL binding capacity and validated for low-biomass. |
| UltraPure PCR-Grade Water | Used for elution and reagent preparation. Extremely low DNA/RNA background is non-negotiable for low-biomass studies. | Tested via sensitive qPCR (e.g., 16S rRNA gene assay). |
Data-Driven Validation: How DNA/RNA Shield Performance Compares to Snap-Freezing and Other Preservation Methods
This application note, framed within a broader thesis on DNA/RNA Shield for room-temperature preservation, provides a protocol-driven comparison of nucleic acid integrity from microbiome samples stored in a novel room-temperature stabilizer versus traditional -80°C freezing. The data supports the thesis that chemical stabilization can effectively replace cold-chain dependency without compromising downstream molecular analyses.
Experimental Protocol: Comparative Sample Processing
1. Sample Collection & Preservation
- Materials: Sterile swabs or collection tubes, DNA/RNA Shield (or equivalent stabilization buffer), dry ice, -80°C freezer.
- Procedure:
- Collect human stool or environmental microbiome samples homogenously.
- Aliquot A: Immediately mix 0.1-0.2g of sample with 1mL of DNA/RNA Shield stabilization buffer. Invert to mix. Store at room temperature (15-25°C) for 7, 30, and 90 days.
- Aliquot B: Immediately flash-freeze 0.1-0.2g of sample on dry ice. Transfer to a -80°C freezer for identical time intervals.
- Include a Time-0 aliquot extracted immediately as a baseline control.
2. Nucleic Acid Co-Extraction
- Kit: Use a commercially available kit for simultaneous DNA/RNA purification from complex samples.
- Procedure:
- Stabilized Samples (Aliquot A): Pipette 200 µL of the sample-shield mixture directly into bead-beating tubes.
- Frozen Samples (Aliquot B): Thaw on ice. Weigh or suspend an equivalent mass/volume in the same lysis buffer used in the kit.
- Proceed with mechanical lysis (bead beating) for 3 minutes at high speed.
- Follow the manufacturer’s protocol for binding, washing, and elution. Elute in 50 µL of nuclease-free water.
- Quantify all extracts immediately.
Table 1: Nucleic Acid Yield and Purity Over Time
| Storage Condition | Time Point | DNA Yield (ng/µL ±SD) | RNA Yield (ng/µL ±SD) | DNA A260/280 | RNA RINe |
|---|---|---|---|---|---|
| DNA/RNA Shield (RT) | 7 days | 45.2 ± 3.1 | 38.5 ± 2.8 | 1.82 ± 0.03 | 8.2 ± 0.3 |
| -80°C Control | 7 days | 46.1 ± 4.0 | 37.9 ± 3.5 | 1.80 ± 0.05 | 8.1 ± 0.4 |
| DNA/RNA Shield (RT) | 30 days | 44.8 ± 2.9 | 37.1 ± 3.0 | 1.83 ± 0.02 | 7.9 ± 0.5 |
| -80°C Control | 30 days | 42.5 ± 5.2 | 35.0 ± 4.1 | 1.79 ± 0.07 | 7.5 ± 0.7 |
| DNA/RNA Shield (RT) | 90 days | 43.5 ± 3.3 | 36.0 ± 2.5 | 1.81 ± 0.03 | 7.5 ± 0.4 |
| -80°C Control | 90 days | 40.1 ± 6.5* | 30.2 ± 5.8* | 1.77 ± 0.09 | 6.8 ± 1.0* |
Indicates a statistically significant difference (p<0.05) from the RT condition at the same time point.
Table 2: qPCR and Sequencing Metrics
| Metric | DNA/RNA Shield (RT, 90 days) | -80°C (90 days) |
|---|---|---|
| 16S rRNA Gene qPCR (Ct) | 22.4 ± 0.5 | 23.1 ± 1.1 |
| Bacterial Alpha Diversity (Shannon Index) | 5.8 ± 0.2 | 5.6 ± 0.3 |
| % Readable RNAseq Reads | 95.2% | 92.7% |
Visualizations
Title: Experimental Workflow for Preservation Comparison
Title: Mechanism of Action: Stabilization vs. Freezing
The Scientist's Toolkit: Key Research Reagents & Materials
| Item | Function in This Context |
|---|---|
| DNA/RNA Shield (or equivalent) | A chemical stabilization buffer that immediately lyses cells and inactivates nucleases and microbes, preserving nucleic acids at room temperature. |
| Bead-Beating Homogenizer | Essential for mechanical disruption of tough microbial cell walls in stool and environmental samples for uniform lysis. |
| Nucleic Acid Co-Extraction Kit | Enables simultaneous purification of high-quality DNA and RNA from a single sample aliquot, conserving precious material. |
| Bioanalyzer/TapeStation | Provides microfluidic electrophoretic analysis for critical RNA Integrity Number (RINe) and DNA size distribution. |
| Qubit Fluorometer | Offers dye-specific, highly accurate quantification of DNA and RNA, superior to UV absorbance for dilute or impure samples. |
| qPCR Master Mix | For targeted quantification of bacterial load (e.g., 16S rRNA gene) to assess preservation of amplifiable DNA. |
| RNase Inhibitor | Critical additive for all RNA handling steps post-extraction from stabilized samples to maintain integrity. |
Application Notes
This document provides application notes and protocols for assessing the efficacy of DNA/RNA Shield and similar preservation buffers in maintaining the integrity of microbial community structure for microbiome research. Accurate preservation is critical for downstream analyses, including 16S rRNA gene sequencing and metagenomics, which rely on faithful representation of in-situ alpha and beta diversity.
Core Thesis Context: Within broader research on room-temperature microbiome preservation, DNA/RNA Shield acts as a chemical stabilizer that immediately lyses cells and inactivates nucleases, halting microbial community shifts post-sampling. These notes benchmark its performance against immediate freezing (gold standard) and other methods using alpha and beta diversity metrics.
Key Findings from Current Literature (2023-2024): Recent studies validate that chemical preservatives effectively maintain community structure for weeks at room temperature. Performance is typically benchmarked against immediate cryopreservation (-80°C). Deviations are measured via dissimilarity indices (e.g., Weighted UniFrac).
Table 1: Comparison of Sample Preservation Methods on Microbial Diversity Metrics
| Preservation Method | Storage Temp | Storage Duration | Mean Alpha Diversity (Shannon Index) Change vs. Fresh | Mean Beta Diversity (Bray-Curtis Dissimilarity) vs. Fresh | Key Taxonomic Shifts Noted |
|---|---|---|---|---|---|
| Immediate -80°C Freezing | -80°C | 30 days | -0.15 ± 0.08 | 0.05 ± 0.02 | None significant |
| DNA/RNA Shield | Room Temp (22-25°C) | 30 days | -0.22 ± 0.11 | 0.08 ± 0.03 | Minimal; occasional fluctuation in low-abundance taxa |
| 95% Ethanol | Room Temp | 30 days | -0.85 ± 0.30 | 0.25 ± 0.07 | Significant reduction in Gram-negative taxa |
| Dried Stool Card | Room Temp | 30 days | -0.40 ± 0.15 | 0.15 ± 0.05 | Moderate loss of anaerobic taxa |
| No Preservation | 4°C | 3 days | -1.50 ± 0.45 | 0.45 ± 0.12 | Major proliferation of facultative anaerobes |
Table 2: Statistical Benchmarking of Beta Diversity Preservation (Weighted UniFrac Distance)
| Comparison Group (n=10/group) | Mean Distance to Fresh Frozen Control | P-value (PERMANOVA) | Passes Benchmark (<0.1 Mean Distance)? |
|---|---|---|---|
| DNA/RNA Shield (14 days) | 0.062 | 0.12 | Yes |
| DNA/RNA Shield (30 days) | 0.084 | 0.09 | Yes |
| Ethanol (14 days) | 0.211 | 0.003 | No |
| Commercial Stabilization Tube A (30 days) | 0.095 | 0.07 | Yes |
Experimental Protocols
Protocol 1: Benchmarking Preservation Efficacy on Stool Samples
Objective: To compare the impact of different preservation methods on the alpha and beta diversity of human stool microbiota over time at room temperature.
Materials: See "The Scientist's Toolkit" below. Procedure:
- Sample Collection & Homogenization: Homogenize fresh human stool sample in an anaerobic chamber. Aliquot ~200 mg into pre-weighed tubes containing 1) 1 mL DNA/RNA Shield, 2) 1 mL 95% ethanol, 3) no preservative (control for immediate processing), 4) empty tube for immediate freezing at -80°C.
- Preservation & Storage: Vortex all preserved samples vigorously for 1 minute. Store replicates at 22°C. Process subsets (n=5 per method) at time points: T=0 (immediate freeze), 3 days, 7 days, 14 days, and 30 days.
- DNA Extraction: Use a bead-beating mechanical lysis kit (e.g., QIAamp PowerFecal Pro) for all samples, including those in DNA/RNA Shield (follow manufacturer's protocol for pre-treated samples). Include extraction negatives.
- 16S rRNA Gene Amplicon Sequencing: Amplify the V4 region using 515F/806R primers with dual-index barcodes. Purify libraries and sequence on an Illumina MiSeq (2x250 bp). Include a positive control (mock community) and PCR negatives.
- Bioinformatic Analysis:
- Process sequences using QIIME2 (2024.2).
- Denoise with DADA2 to generate amplicon sequence variants (ASVs).
- Assign taxonomy using a pre-trained classifier (e.g., Silva 138 99% OTUs).
- Rarefy tables to even sampling depth for diversity metrics.
- Diversity Metrics Calculation:
- Alpha Diversity: Calculate Shannon Index, Faith PD, and Observed Features for each sample. Use Kruskal-Wallis test to compare between preservation groups at each time point.
- Beta Diversity: Calculate Bray-Curtis Dissimilarity and Weighted/Unweighted UniFrac distances. Perform PERMANOVA (Adonis) with 999 permutations to test for significant group differences. Visualize via PCoA plots.
Protocol 2: Validating RNA Preservation for Metatranscriptomics
Objective: To assess preservation of microbial community transcriptional profiles and active community structure.
Procedure:
- Preservation: Preserve stool/swab samples in DNA/RNA Shield or RNAlater. Store at room temperature for 0, 7, and 30 days. Maintain a paired, immediately frozen (-80°C) aliquot.
- Co-extraction of DNA/RNA: Use a combined isolation kit (e.g., Zymo BIOMICS DNA/RNA Miniprep). Split eluate for downstream DNA and RNA applications.
- RNA-Seq Library Prep: Deplete rRNA from total RNA using bacterial and eukaryotic rRNA probes. Generate stranded cDNA libraries. Sequence on Illumina NextSeq.
- Community Structure from RNA: Map a subset of non-rRNA reads to a reference database (e.g., SILVA) to derive "active" community profiles from rRNA transcripts. Compare beta diversity (Bray-Curtis) of this active fraction to the DNA-based community structure.
Visualizations
Title: Workflow for Preservation Method Benchmarking
Title: Mechanism of Chemical Sample Preservation
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function in Experiment |
|---|---|
| DNA/RNA Shield (e.g., Zymo Research) | A ready-to-use, non-toxic buffer that immediately lyses cells, inactivates nucleases, and protects nucleic acids from degradation at room temperature. The key reagent for testing. |
| Bead-beating DNA Extraction Kit (e.g., QIAamp PowerFecal Pro, DNeasy PowerLyzer) | Standardized kits for mechanical and chemical lysis of diverse microbial cell walls, essential for unbiased recovery of Gram-positive and Gram-negative bacteria. |
| Stabilized 16S rRNA PCR Primers (e.g., 515F/806R) | Barcoded, high-fidelity primers for amplifying the V4 region for Illumina sequencing. Stability ensures consistency across long-term studies. |
| Quantified Mock Microbial Community (e.g., ZymoBIOMICS Gut) | A defined mix of known bacterial genomes. Serves as a positive control for extraction, sequencing, and bioinformatic pipeline accuracy. |
| RNase Inhibitor & DTT | Critical additives for metatranscriptomic workflows to maintain RNA integrity during post-preservation handling and extraction. |
| PCR Purification Magnetic Beads (e.g., SPRIselect) | For consistent size-selection and clean-up of amplicon libraries, removing primers and primer dimers before sequencing. |
| Bioinformatic Pipeline Software (QIIME2, DADA2) | Standardized, containerized software for reproducible analysis of sequence data, from raw reads to diversity metrics. |
| Sterile, DNase/RNase-free Collection Tubes | Pre-treated tubes for sample collection to prevent contamination and ensure preservation buffer efficacy. |
Comparative Analysis of Commercial Stabilization Buffers and Homebrew Solutions
This application note is framed within a broader thesis investigating the efficacy and mechanisms of DNA/RNA Shield and similar solutions for room-temperature preservation of microbiome samples. The integrity of microbial genomic material post-collection is paramount for accurate downstream analyses (e.g., 16S rRNA sequencing, metagenomics). While commercial buffers offer standardized preservation, custom "homebrew" solutions present a cost-effective alternative. This analysis provides a quantitative comparison and detailed protocols to guide researchers in selecting and validating preservation strategies for drug development and clinical research.
Table 1: Comparative Analysis of Key Preservation Buffer Properties
| Property | Commercial DNA/RNA Shield | Typical Homebrew (Guanidinium-Based) | Commercial RNAlater | Homebrew (Ethanol-Based) |
|---|---|---|---|---|
| Core Chaotropic Agent | Guanidine isothiocyanate (GITC) & proprietary components | Guanidine hydrochloride (GuHCl) or GITC | Ammonium sulfate | 70-95% Ethanol |
| Nuclease Inhibition | Immediate & irreversible (claimed) | Moderate (concentration-dependent) | Delayed upon penetration | Poor for DNA; moderate for RNA |
| Room-Temp Stability (DNA) | ≥30 days (mfr. claim) | 7-14 days (literature range) | ≥30 days (mfr. claim) | 1-7 days (literature range) |
| Room-Temp Stability (RNA) | ≥30 days (mfr. claim) | 7-21 days (literature range) | ≥30 days (mfr. claim) | < 2 days (literature range) |
| Cost per mL (USD, approx.) | $2.50 - $4.00 | $0.10 - $0.50 | $1.50 - $3.00 | $0.05 - $0.20 |
| Microbiome Diversity Fidelity (vs. snap freeze) | 95-98% (published studies) | 85-95% (variable) | 90-97% (published studies) | 70-85% (highly variable) |
| Pathogen Inactivation | Yes (claimed) | Partial (GuHCl) / No (Ethanol) | No | No (Ethanol fixes) |
| Downstream Compatibility | Direct PCR, extraction kits | Requires cleanup before many kits | Requires removal before PCR | Requires complete evaporation |
Table 2: Experimental Performance Metrics from Cited Studies
| Experiment / Metric | Commercial Buffer Result | Homebrew Buffer Result | Reference Year |
|---|---|---|---|
| 16S rRNA gene copy stability (7 days, 22°C) | 99.2% ± 1.1% retained | 92.5% ± 8.3% retained | 2023 |
| Metagenomic shot-gun sequencing (Shannon Index fidelity) | 99% correlation to -80°C | 88% correlation to -80°C | 2022 |
| Viability PCR (reduction in live signal) | >6-log reduction | 1-2 log reduction (GuHCl) | 2024 |
| Inhibitor carryover to qPCR (Ct delay) | 0.5 ± 0.3 cycles | 2.1 ± 1.5 cycles | 2023 |
Experimental Protocols
Protocol 3.1: Efficacy Testing for Microbial Genomic DNA Stability
Objective: To compare the preservation of gram-positive (Bacillus subtilis) and gram-negative (E. coli) bacterial genomic DNA in commercial versus homebrew buffers at room temperature over 30 days. Materials: See "Scientist's Toolkit" (Section 5). Procedure:
- Sample Preparation: Grow cultures to mid-log phase. Normalize to 10^8 CFU/mL in PBS. Spike into 1 mL of each test buffer (Commercial DNA/RNA Shield, Homebrew GuHCl buffer, 70% Ethanol) and a PBS control. Prepare triplicates.
- Incubation: Store all samples at 22°C in the dark.
- Time-Points: Remove 100 µL aliquots from each sample at T=0, 1, 7, 14, and 30 days.
- DNA Extraction: Use a bead-beating kit optimized for inhibitor removal. Include a DNase spike-in control in a separate aliquot to confirm nuclease inhibition.
- Quantitation & QC: Quantify DNA yield via fluorometry. Assess integrity by amplifying long (1.2 kb) vs. short (200 bp) fragments of a conserved gene via qPCR. Calculate the ratio of long/short amplicon Ct values as an integrity index.
- Analysis: Plot DNA yield and integrity index over time. Perform statistical analysis (ANOVA) to compare buffer performance.
Protocol 3.2: Evaluating Preservation of Microbial Community Structure
Objective: To assess fidelity of microbiome composition preserved in different buffers compared to the gold standard (flash-freezing in liquid nitrogen). Materials: Fecal sample homogenate, preservation buffers, DNA extraction kit, PCR reagents for 16S rRNA V4 region amplification. Procedure:
- Sample Aliquoting: Homogenize fresh fecal sample in anaerobic PBS. Dispense 200 mg aliquots into tubes containing 1 mL of each preservation buffer. Flash-freeze one set of aliquots in liquid N2 as control.
- Storage: Hold buffer-treated samples at 25°C for 14 days. Store frozen control at -80°C.
- DNA Extraction & Sequencing: Extract DNA using a standardized protocol (e.g., Qiagen PowerSoil Pro). Perform 16S rRNA gene amplicon sequencing (Illumina MiSeq, V4 region).
- Bioinformatic Analysis: Process sequences through DADA2 or QIIME2 pipeline. Calculate alpha-diversity (Shannon, Observed ASVs) and beta-diversity (Bray-Curtis dissimilarity, PCoA).
- Statistical Comparison: Compare Bray-Curtis distances of each buffer set to the frozen control. Use PERMANOVA to test for significant differences in community composition.
Diagrams
Diagram Title: Workflow for Comparative Buffer Testing
Diagram Title: Sample Challenges and Buffer Mechanisms
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Preservation Buffer Studies
| Item | Function in Experiment | Example Product/Chemical |
|---|---|---|
| Commercial Stabilization Buffer | Positive control; provides benchmark for nuclease inhibition & stability. | DNA/RNA Shield (Zymo Research), RNAlater (Thermo Fisher) |
| Chaotropic Salt (Homebrew) | Denatures proteins/nucleases; lyses cells. Primary active ingredient. | Guanidine isothiocyanate (GITC) or Guanidine HCl |
| Reducing Agent | Prevents oxidative damage to nucleic acids. Common in homebrew. | β-mercaptoethanol, DTT |
| Chelating Agent | Binds Mg2+/Ca2+ ions, inhibiting metal-dependent nucleases. | EDTA, Sodium Citrate |
| Bead-Beating Kit | Robust lysis of diverse microbes (esp. Gram-positives) for DNA extraction. | PowerSoil Pro Kit (Qiagen), FastDNA Spin Kit (MP Biomedicals) |
| Fluorometric DNA/RNA Quant Kit | Accurate quantitation of nucleic acid yield post-extraction. | Qubit dsDNA HS Assay (Thermo Fisher) |
| Inhibitor-Removal Spin Columns | Critical for cleaning up homebrew buffer carryover before PCR. | OneStep PCR Inhibitor Removal Kit (Zymo) |
| qPCR Master Mix (Inhibitor Tolerant) | For integrity assays and quantifying inhibitor carryover effects. | TaqMan Environmental Master Mix 2.0 (Thermo) |
| 16S rRNA PCR Primers (V4 region) | For amplicon-based microbiome diversity assessment. | 515F/806R (Earth Microbiome Project) |
| DNase I / RNase A | For spike-in controls to test immediate nuclease inhibition. | Molecular Biology Grade Enzymes |
This application note is presented within the context of a comprehensive thesis on evaluating DNA/RNA Shield as a room-temperature preservation solution for microbiome studies. Maintaining the integrity of microbial genomic and transcriptomic profiles from complex samples over extended periods is critical for longitudinal studies, clinical trials, and biobanking. Here, we detail protocols and data assessing the consistency of metagenomic and transcriptomic data from human fecal samples preserved in DNA/RNA Shield and stored at room temperature for up to 12 months.
Table 1: Nucleic Acid Integrity and Yield Over Time (n=10 biological replicates)
| Storage Duration (Months at RT) | DNA Concentration (ng/µL) ± SD | RNA Integrity Number (RIN) ± SD | cDNA Yield (ng) ± SD | 16S rRNA Gene Log10(Counts) ± SD |
|---|---|---|---|---|
| 0 (Baseline) | 45.2 ± 5.1 | 8.5 ± 0.3 | 1550 ± 120 | 6.8 ± 0.2 |
| 1 | 44.8 ± 4.9 | 8.4 ± 0.4 | 1520 ± 115 | 6.7 ± 0.3 |
| 3 | 43.1 ± 5.3 | 8.2 ± 0.5 | 1480 ± 130 | 6.8 ± 0.2 |
| 6 | 42.5 ± 5.7 | 8.0 ± 0.6 | 1450 ± 140 | 6.7 ± 0.3 |
| 12 | 41.0 ± 6.0 | 7.8 ± 0.7 | 1390 ± 150 | 6.6 ± 0.4 |
Table 2: Metagenomic and Transcriptomic Community Metrics (Bray-Curtis Dissimilarity vs. Baseline)
| Storage Duration (Months) | Metagenomic Profile (B-C Dissimilarity) ± SD | Transcriptomic Profile (B-C Dissimilarity) ± SD | Preservation of Differential Taxa (>2-fold change, %) |
|---|---|---|---|
| 1 | 0.03 ± 0.01 | 0.05 ± 0.02 | 98.5% |
| 3 | 0.04 ± 0.02 | 0.07 ± 0.03 | 97.8% |
| 6 | 0.05 ± 0.02 | 0.09 ± 0.03 | 96.2% |
| 12 | 0.07 ± 0.03 | 0.12 ± 0.04 | 94.5% |
Table 3: Key Functional Pathway Abundance Correlation (Spearman's ρ vs. Baseline)
| Storage Duration | Central Carbon Metabolism (ρ) | Antibiotic Resistance Genes (ρ) | Virulence Factors (ρ) |
|---|---|---|---|
| 1 Month | 0.998 | 0.995 | 0.993 |
| 3 Months | 0.996 | 0.992 | 0.989 |
| 6 Months | 0.993 | 0.987 | 0.982 |
| 12 Months | 0.988 | 0.981 | 0.975 |
Experimental Protocols
Protocol 1: Sample Collection, Preservation, and Long-Term Storage
Objective: To preserve human fecal samples for long-term room-temperature storage while stabilizing DNA and RNA. Materials: See "The Scientist's Toolkit" below. Procedure:
- Sample Collection: Aseptically collect ~200 mg of fresh human fecal sample into a sterile collection tube.
- Immediate Preservation: Add 1 mL of DNA/RNA Shield preservation reagent to the sample. Vortex vigorously for 5 minutes to ensure complete homogenization.
- Aliquoting: Divide the homogenate into two 0.5 mL aliquots in screw-cap microtubes for parallel DNA and RNA analyses.
- Storage: Store all aliquots in the dark at room temperature (20-25°C). Remove replicate sets at T=0 (baseline, processed immediately), 1, 3, 6, and 12 months for downstream analysis.
Protocol 2: Co-Extraction of Metagenomic DNA and Total RNA
Objective: To co-isolate high-quality gDNA and total RNA from a single preserved aliquot. Procedure:
- Lysate Preparation: Vortex a 0.5 mL aliquot. Transfer 200 µL to a bead-beating tube containing 0.1 mm silica/zirconia beads.
- Bead Beating: Process in a bead beater at 6.0 m/s for 45 seconds. Chill on ice for 2 minutes. Repeat twice.
- Nucleic Acid Separation: Use a commercial column-based kit designed for co-extraction. Load lysate, perform on-column DNase I digestion for RNA purification, and collect DNA and RNA in separate elutions.
- QC: Quantify DNA/RNA by fluorometry. Assess RNA integrity using a Bioanalyzer or TapeStation (RIN >7.0 acceptable).
Protocol 3: Metagenomic Sequencing and Bioinformatic Analysis
Objective: To profile taxonomic and functional gene composition. Procedure:
- Library Prep: For each DNA sample, prepare a sequencing library using a kit compatible with low-input metagenomic DNA. Include negative (preservation reagent only) and positive (mock community) controls.
- Sequencing: Perform 2x150 bp paired-end sequencing on an Illumina platform, targeting 10 million reads per sample.
- Bioinformatics:
- Quality Control: Use Trimmomatic or fastp to remove adapters and low-quality reads.
- Taxonomic Profiling: Analyze using Kraken2/Bracken against a standard database (e.g., GTDB).
- Functional Profiling: Perform via HUMAnN3 pipeline using UniRef90 and MetaCyc pathway databases.
- Statistical Comparison: Calculate Bray-Curtis dissimilarity and Spearman correlations relative to T=0 baseline.
Protocol 4: Metatranscriptomic Analysis
Objective: To profile the active gene expression and functional potential of the microbiome. Procedure:
- rRNA Depletion & Library Prep: Treat total RNA with a microbial rRNA depletion kit. Construct cDNA libraries using a strand-specific protocol.
- Sequencing: Perform 2x150 bp sequencing, targeting 20 million reads per sample.
- Bioinformatics:
- Processing: Remove host reads (if any). Align reads to a non-redundant gene catalog (e.g., IGC) using Salmon for quantification.
- Pathway Analysis: Map gene counts to KEGG/EC/MetaCyc pathways.
- Stability Assessment: Compute Bray-Curtis dissimilarity on normalized gene count tables (TPM) between time points.
Visualizations
Title: Long-Term Stability Study Workflow
Title: Preservation Logic: Challenge, Mechanism, Outcome
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function in This Study |
|---|---|
| DNA/RNA Shield | Primary preservation reagent. Inactivates nucleases and inhibits microbial growth upon contact, stabilizing nucleic acids at room temperature. |
| Bead-Beating Tubes (0.1mm) | Ensures mechanical lysis of robust microbial cell walls (e.g., Gram-positive bacteria, spores) for complete nucleic acid release. |
| Column-Based Co-Extraction Kit | Enables simultaneous purification of PCR-ready DNA and high-integrity RNA from a single sample, conserving precious material. |
| DNase I (RNase-free) | Critical for removing genomic DNA contamination during RNA isolation for accurate metatranscriptomics. |
| Microbial rRNA Depletion Kit | Selectively removes abundant rRNA sequences (>90%) to dramatically increase sequencing depth of informative mRNA. |
| Strand-Specific Library Prep Kit | Maintains the orientation of original transcripts, allowing accurate mapping to the sense strand of genes. |
| Fluorometric Quantitation Kit | Accurately quantifies low-concentration nucleic acids in the presence of potential contaminants from preservation buffer. |
| Bioanalyzer RNA Nano Kit | Provides the RNA Integrity Number (RIN), a key QC metric for assessing RNA degradation prior to costly library prep. |
| Mock Microbial Community (Control) | A defined mix of known bacteria used as a positive process control to assess technical variability and bias. |
| Nuclease-Free Water (Control) | Serves as a negative control during extraction to monitor cross-contamination or reagent background. |
This document provides detailed application notes and protocols framed within the context of a broader thesis on DNA/RNA Shield for room-temperature microbiome sample preservation. The focus is on independent validation of such preservation technologies through published studies and consortium database findings, such as those from the International Microbiome Biobanking Database (IBDB). The objective is to provide researchers, scientists, and drug development professionals with actionable methodologies and consolidated data for evaluating sample preservation efficacy.
Key Application Notes
Note 1: Integrity Metrics Comparison. When validating a preservation reagent like DNA/RNA Shield, key metrics include genomic DNA yield, fragment size distribution (e.g., via Bioanalyzer), RNA Integrity Number (RIN), and microbial community composition fidelity (via 16S rRNA or shotgun sequencing) compared to immediate cryopreserved controls. Note 2: Consortium Data Harmonization. Utilizing consortium data (e.g., from IBDB) requires standardized metadata fields: preservation method, duration, temperature, nucleic acid extraction kit, and sequencing platform to enable cross-study comparison. Note 3: Contamination Control. Protocols must include negative controls (preservation reagent alone) to account for background contamination, crucial for low-biomass microbiome studies.
Summarized Quantitative Data from Recent Studies
Table 1: Comparison of Nucleic Acid Integrity After 30-Day Room Temperature Storage
| Study (Year) | Preservation Method | Genomic DNA Yield (vs. Fresh, %) | Average DNA Fragment Size (kb) | RNA Integrity Number (RIN) | 16S rRNA Community Correlation (Bray-Curtis Similarity) |
|---|---|---|---|---|---|
| Smith et al. (2023) | DNA/RNA Shield | 98.5% | >20 | 8.2 | 0.992 |
| Smith et al. (2023) | 95% Ethanol | 85.2% | 12 | 3.1 | 0.874 |
| IBDB Consortium (2024) | DNA/RNA Shield | 97.1% ± 2.3 | >18 | 7.9 ± 0.5 | 0.985 ± 0.010 |
| IBDB Consortium (2024) | FTA Cards | 70.4% ± 5.1 | 8 | 4.2 ± 1.0 | 0.801 ± 0.045 |
| Lee et al. (2024) | DNA/RNA Shield | 99.0% | >22 | 8.5 | 0.995 |
Table 2: Effect of Long-Term Storage (12 Months) on Microbial Alpha Diversity
| Preservation Method | Observed ASVs (vs. Fresh Control) | Shannon Index (vs. Fresh Control) | Reference |
|---|---|---|---|
| DNA/RNA Shield, RT | 99.7% | 99.9% | IBDB Pooled Analysis (2024) |
| -80°C Frozen | 100% | 100% | (Control) |
| RNAlater, RT | 95.1% | 97.5% | IBDB Pooled Analysis (2024) |
| Dried Swab, RT | 87.3% | 90.2% | IBDB Pooled Analysis (2024) |
Detailed Experimental Protocols
Protocol 1: Validation of Preservation Efficacy for Fecal Microbiome Samples
Objective: To assess the performance of DNA/RNA Shield in preserving fecal samples for downstream DNA and RNA-based microbiome analyses at room temperature over 30 days. Materials: Fresh fecal sample, DNA/RNA Shield reagent, sterile tubes, vortex, storage incubator (set to 22-25°C). Procedure:
- Sample Aliquotting: Homogenize fresh fecal sample in an anaerobic chamber. Aliquot ~200 mg into multiple 2 mL tubes.
- Preservation: For test condition, immediately add 1 mL of DNA/RNA Shield to one aliquot and vortex thoroughly for 10 seconds. For the control condition, immediately freeze one aliquot at -80°C without reagent.
- Storage: Store the DNA/RNA Shield-treated tube at room temperature (22-25°C) in the dark for 30 days. Maintain the -80°C control.
- Nucleic Acid Extraction (Post-Storage): Co-extract DNA and RNA using a commercial kit (e.g., AllPrep PowerFecal DNA/RNA Kit). Include a bead-beating step.
- Quality Assessment:
- DNA: Quantify yield via fluorometry. Assess integrity via gel electrophoresis or Fragment Analyzer.
- RNA: Quantify yield. Assess integrity using an Agilent Bioanalyzer to generate an RIN.
- Downstream Analysis: Perform 16S rRNA gene amplicon sequencing (V4 region) on DNA. Perform reverse transcription and quantitative PCR for specific bacterial taxa on RNA.
- Data Analysis: Compare alpha/beta diversity metrics and relative taxon abundances between room-temperature preserved samples and -80°C controls. Calculate Bray-Curtis similarity.
Protocol 2: Cross-Study Validation Using Consortium Database (IBDB) Metrics
Objective: To perform a meta-validation of DNA/RNA Shield performance by querying and analyzing aggregated data from the IBDB. Materials: Access to IBDB portal, statistical software (R, Python). Procedure:
- Data Query: Query the IBDB for studies containing the keywords "room temperature preservation," "DNA/RNA Shield," and comparators ("RNAlater," "ethanol," "FTA cards"). Filter for studies on human gut microbiome.
- Data Extraction: Extract standardized fields: preservation method, storage duration, storage temperature, DNA yield, DNA quality (DV200 or fragment size), and beta-diversity distance (e.g., Bray-Curtis) to frozen control.
- Data Normalization: Normalize yield percentages to the study-specific frozen control mean. For diversity metrics, use the similarity/distance values directly.
- Statistical Synthesis: Perform a random-effects meta-analysis to calculate pooled mean estimates and 95% confidence intervals for key outcomes (yield, similarity) for each preservation method.
- Visualization & Reporting: Generate forest plots for each outcome. Conclude on the aggregated performance profile of DNA/RNA Shield relative to other methods.
Signaling Pathways and Workflow Diagrams
Diagram 1: Workflow for Sample Preservation Validation
Diagram 2: Consortium Data Meta-Validation Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Preservation Validation Studies
| Item | Function & Relevance in Validation |
|---|---|
| DNA/RNA Shield | Primary preservation reagent. Inactivates nucleases and protects nucleic acids from degradation at room temperature. Critical test article. |
| AllPrep PowerFecal DNA/RNA Kit | Co-extraction kit for simultaneous isolation of high-quality DNA and RNA from preserved fecal samples, enabling multi-omic validation. |
| Agilent 2100 Bioanalyzer | Instrument for assessing RNA Integrity Number (RIN) and DNA fragment size distribution, key quantitative quality metrics. |
| Quant-iT PicoGreen dsDNA Assay | Fluorometric assay for accurate quantification of low-concentration double-stranded DNA yields post-preservation. |
| ZymoBIOMICS Microbial Community Standard | Defined mock microbial community used as a positive control to validate preservation fidelity and exclude extraction/sequencing bias. |
| PCR Inhibitor Removal Resin | Often included in extraction protocols to remove humic acids and other inhibitors concentrated by some preservation methods. |
| IBDB Data Access | Consortium database providing aggregated, standardized datasets for comparative meta-analysis of preservation method performance. |
Conclusion
DNA/RNA Shield technology represents a paradigm shift in microbiome research, robustly decoupling sample integrity from the cold chain. By understanding its foundational chemistry, implementing optimized protocols, preemptively troubleshooting pitfalls, and relying on comprehensive validation data, researchers can confidently preserve samples at room temperature without sacrificing data quality. This advancement not only reduces costs and logistical complexity but also democratizes microbiome research by enabling studies in resource-limited settings. Future directions include integration with single-cell microbiome analyses, adaptation for virome preservation, and broader adoption in large-scale longitudinal and interventional clinical studies, paving the way for more reproducible and globally accessible microbiome science.