Shotgun Metagenomic Sequencing: A Comprehensive Guide to Microbiome Functional Analysis for Biomedical Research
This article provides a comprehensive overview of shotgun metagenomic sequencing for microbiome functional analysis, tailored for researchers, scientists, and drug development professionals.
Shotgun Metagenomic Sequencing: A Comprehensive Guide to Microbiome Functional Analysis for Biomedical Research
Abstract
This article provides a comprehensive overview of shotgun metagenomic sequencing for microbiome functional analysis, tailored for researchers, scientists, and drug development professionals. It covers foundational principles, from distinguishing shotgun sequencing from 16S rRNA methods to exploring its ability to characterize unculturable microbes and reveal functional genetic potential. The piece details methodological workflows and applications in pharmaceutical development, including antimicrobial resistance tracking and therapeutic discovery. It also addresses critical troubleshooting considerations for complex samples and data analysis, and concludes with validation strategies and comparative analyses against other microbiome profiling techniques. This resource serves as a practical guide for leveraging metagenomic insights in clinical and biotechnological applications.
Unlocking Microbial Dark Matter: Foundational Principles of Shotgun Metagenomics
What is Shotgun Metagenomic Sequencing? Definition and Core Principles
Definition and Core Principles
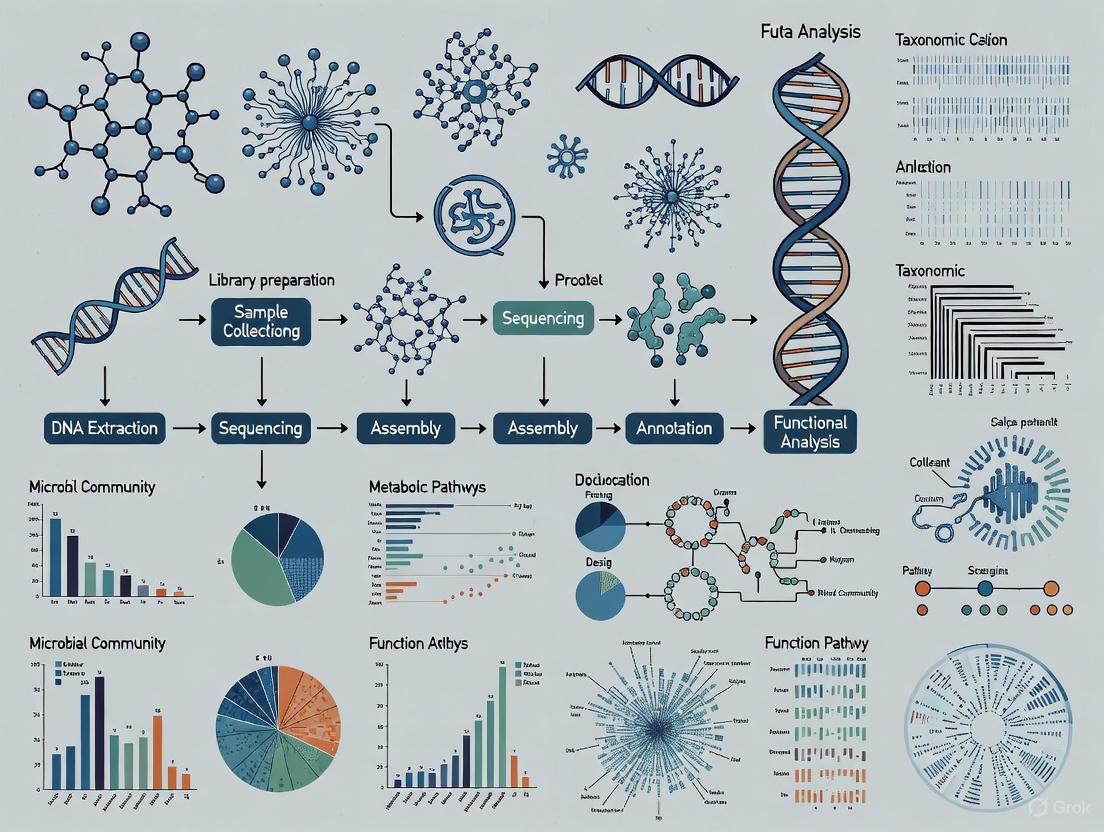
Shotgun metagenomic sequencing is a next-generation sequencing (NGS) method that involves comprehensively sampling and sequencing all genes from all organisms present in a given complex sample [1]. The core principle of this technique lies in its non-targeted approach: instead of amplifying specific marker genes, all genomic DNA extracted from a sample—whether from microbes, viruses, or other biological entities—is randomly fragmented into small pieces, much like a shotgun would break something into pieces [2]. These small DNA fragments are then sequenced in parallel, generating millions of reads that are subsequently analyzed and stitched back together using sophisticated bioinformatics tools [2].
The fundamental workflow can be broken down into several key stages, as illustrated below:
This approach enables researchers to simultaneously evaluate bacterial diversity and detect the abundance of microbes in various environments, including the study of unculturable microorganisms that are otherwise difficult or impossible to analyze [1]. Unlike targeted methods such as 16S rRNA gene sequencing, shotgun metagenomics provides a complete picture of the microbial community by capturing genetic material from all domains of life—bacteria, archaea, fungi, and viruses—while also enabling researchers to elucidate the functional potential of these communities [3] [2] [4].
Key Advantages and Comparative Analysis
Shotgun metagenomic sequencing offers several distinct advantages over targeted sequencing approaches, which are summarized in the table below.
Table 1: Comparative analysis of shotgun metagenomic sequencing versus 16S rRNA amplicon sequencing
| Feature | Shotgun Metagenomic Sequencing | 16S rRNA Amplicon Sequencing |
|---|---|---|
| Scope of Detection | All microorganisms (bacteria, archaea, fungi, viruses) [2] [4] | Primarily bacteria and archaea only [4] |
| Taxonomic Resolution | Species to strain level [2] | Typically genus level, sometimes species [5] |
| Functional Insights | Direct assessment of functional genes and metabolic pathways [3] [2] | Limited to phylogenetic inference (e.g., PICRUSt) [6] |
| PCR Bias | Minimal (no targeted amplification) [2] | Significant (primers target specific regions) [5] |
| Reference Database Dependence | Dependent on genomic databases [2] [5] | Dependent on 16S-specific databases (e.g., SILVA, Greengenes) [5] |
| Cost Considerations | Higher cost, especially for deep sequencing [2] | More cost-effective [5] |
| Bioinformatic Complexity | High computational requirements [2] | Less computationally intensive [5] |
| Host DNA Contamination | More challenging when host DNA overwhelms microbial DNA [2] [7] | Less affected due to targeted amplification |
The higher resolution of shotgun metagenomics allows for species to strain-level discrimination, a significant advantage over 16S sequencing which typically classifies groups to the genus or species level [2]. Additionally, since there is no PCR amplification step targeting specific regions, shotgun sequencing avoids primer bias, copy-number bias, PCR artifacts, and chimeras that can complicate amplicon-based approaches [2].
Experimental Protocol: A Detailed Workflow
Sample Collection and Preservation
The initial step in any shotgun metagenomics study involves careful sample collection and preservation. Sample types can range from human fecal samples to environmental samples such as soil, water, or air [2]. Three critical factors must be considered:
- Sterility: Sample containers must be sterile to prevent contamination from external microbes [2].
- Temperature: To preserve microbial integrity, samples should be frozen immediately after collection at -20°C or -80°C, or snap-frozen in liquid nitrogen. Freeze-thaw cycles should be avoided as they compromise sample integrity [2].
- Time: Samples should be frozen as quickly as possible after collection. When immediate freezing isn't feasible, temporary storage at 4°C or use of preservation buffers may be suitable alternatives [2].
For human microbiome studies, particularly those involving stool samples, consistency in collection timing and method is crucial for reproducible results.
DNA Extraction
DNA extraction is performed using commercial kits that employ a combination of chemical and physical methods to separate DNA from other cellular components [2]. The key steps include:
- Lysis: Breaking open cells through chemical processes (enzymes) and mechanical processes (vigorous shaking/mixing) to release DNA [2].
- Precipitation: Separating DNA from other cell contents using salt solutions and alcohol [2].
- Purification: Washing the precipitated DNA to remove impurities and resuspending it in an aqueous solution [2].
For challenging samples containing hard-to-lyse structures (e.g., spores) or inhibitors (e.g., humic acids in soil), additional enzymatic or physical treatment steps may be necessary [2]. The choice of DNA extraction kit significantly influences the observed microbial community structure and affects inter-study comparisons.
Library Preparation and Sequencing
Library preparation involves preparing the extracted DNA for sequencing through the following steps:
- DNA Fragmentation: Mechanical or enzymatic methods break DNA into short fragments suitable for sequencing [2].
- Adapter Ligation: Molecular 'barcodes' or index adapters are ligated to DNA fragments to enable sample multiplexing and identification after sequencing [2].
- Library Cleanup: Size selection and purification ensure the DNA library is optimized for sequencing [2].
Sequencing is typically performed using high-throughput platforms such as Illumina, which offers short read lengths (150-300 bp) with high accuracy, or long-read technologies like Oxford Nanopore and PacBio that facilitate assembly of complex genomic regions [3]. The selection of sequencing platform depends on the specific research goals, required read length, and throughput needs [2].
Bioinformatics Analysis
Bioinformatic analysis represents the most computationally intensive phase of shotgun metagenomics. Two primary analytical approaches are employed:
- Read-Based Analysis: Sequencing reads are directly compared to reference databases of microbial marker genes using tools such as Kraken, MetaPhlAn, and HUMAnN [2]. This approach requires less sequencing coverage but is limited by database completeness.
- Metagenome Assembly: Sequencing reads are assembled into partial or complete microbial genomes using tools like MEGAHIT or MetaSPAdes [3] [2]. This approach enables discovery of novel species and strains but demands greater sequencing depth and computational resources.
Table 2: Essential bioinformatics tools for shotgun metagenomic data analysis
| Analysis Step | Tool Examples | Primary Function |
|---|---|---|
| Quality Control | Trimmomatic, Fastp, MultiQC [3] | Remove low-quality reads and sequencing artifacts |
| Host DNA Removal | KneadData, Bowtie2 [8] | Filter out host-derived sequences |
| Metagenome Assembly | MEGAHIT, MetaSPAdes, MetaVelvet [3] | Assemble sequencing reads into contigs |
| Taxonomic Profiling | Kraken, MetaPhlAn, SHOGUN, Woltka [6] [2] | Classify reads taxonomically |
| Functional Annotation | HUMAnN, KEGG, TaxonFinder [3] | Identify functional genes and metabolic pathways |
Recent computational advances such as "sequential co-assembly" have demonstrated significant reductions in assembly time and memory requirements, making metagenomic analysis more accessible in resource-constrained settings [9].
Applications in Microbial Research
Shotgun metagenomic sequencing has enabled groundbreaking research across multiple disciplines:
Human Health and Disease
In biomedical research, shotgun metagenomics has been instrumental in linking microbial communities to human health and disease. Studies of pediatric ulcerative colitis have revealed that affected individuals harbor a dysbiotic and less diverse gut microbial population with distinct differences from healthy children [8]. Similar approaches in colorectal cancer research have identified microbial signatures associated with disease progression, including enrichment of species such as Parvimonas micra and various Fusobacterium species [5]. The predictive power of microbial profiling for disease status has shown remarkable accuracy, with area under the receiver operating characteristic curve (AUROC) values reaching 0.90 in some studies [8].
Environmental Monitoring
In environmental science, shotgun metagenomics provides unprecedented insights into microbial ecosystems. Applications include:
- Soil Microbiome Analysis: Assessing microbial diversity in relation to soil health and fertility, with studies demonstrating that high-fertility soils exhibit greater bacterial diversity, particularly among nitrogen-cycling groups essential for ecosystem functioning [3].
- Water Quality Monitoring: Identifying microbial contaminants and understanding microbial community dynamics in aquatic ecosystems [3].
- Airborne Pathogen Detection: Comprehensive analysis of airborne microbial communities in urban environments to monitor potential pathogens and assess public health risks [3].
Industrial and Food Microbiology
Industrial applications leverage shotgun metagenomics to identify and classify microorganisms in food products, fermented foods, and industrial processes. This enables quality control, pathogen detection, and optimization of biotechnological processes [2].
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Essential research reagents and kits for shotgun metagenomic sequencing
| Reagent/Kits | Manufacturer/Example | Function |
|---|---|---|
| DNA Extraction Kits | QIAamp Powerfecal DNA Kit [8], NucleoSpin Soil Kit [5] | Extraction of high-quality DNA from complex samples |
| Library Preparation Kits | Nextera XT DNA Library Preparation Kit [8] | Preparation of sequencing libraries from extracted DNA |
| Sequenceing Platforms | Illumina NextSeq500 [8], HiSeq Series [3], Oxford Nanopore [3] | High-throughput DNA sequencing |
| Host DNA Removal | KneadData [8] | Bioinformatic filtering of host-derived sequences |
| Positive Control Standards | Commercially available microbial communities | Quality control and protocol validation |
| DBCO-NHCO-PEG4-amine | DBCO-NHCO-PEG4-amine, CAS:1255942-08-5, MF:C29H37N3O6, MW:523.6 g/mol | Chemical Reagent |
| DBCO-PEG4-NHS ester | DBCO-PEG4-NHS ester, MF:C34H39N3O10, MW:649.7 g/mol | Chemical Reagent |
Technical Considerations and Methodological Optimization
Sequencing Depth Considerations
Sequencing depth—the number of sequencing reads aligning to a reference region in a genome—critically impacts data quality. Greater sequencing depth provides stronger evidence for the presence of organisms, particularly low-abundance community members [1]. While traditional shotgun sequencing requires millions of reads per sample, "shallow shotgun sequencing" has emerged as a cost-effective alternative, providing adequate discriminatory power with as few as 500,000 reads per sample while enabling higher discriminatory and reproducible results compared to 16S sequencing [1] [6].
Contamination and Quality Control
As shotgun sequencing detects all genomic DNA in a sample, there is an increased risk of sequencing DNA from unwanted, non-microbial sources. For example, human-associated microbiome studies may generate a high proportion of human reads, with only a small fraction deriving from microbial DNA [2]. Both experimental and computational strategies must be employed to address this challenge, including:
- Implementation of negative controls to identify potential contaminants [2]
- Bioinformatics tools for host DNA removal (e.g., KneadData, Bowtie2 against host genomes) [8]
- Careful sample processing protocols to minimize external contamination [2]
The following diagram illustrates the complete shotgun metagenomic sequencing workflow with key decision points:
Shotgun metagenomic sequencing represents a powerful and transformative approach for studying complex microbial communities without the limitations of cultivation or targeted amplification. By providing comprehensive insights into both taxonomic composition and functional potential, this method has advanced our understanding of microbial ecology in diverse environments, from the human body to ecosystems worldwide. While the approach demands significant computational resources and bioinformatics expertise, ongoing methodological improvements—such as shallow sequencing and efficient assembly algorithms—continue to enhance its accessibility. As reference databases expand and analytical tools mature, shotgun metagenomics will undoubtedly remain a cornerstone technique in microbial research, enabling new discoveries and applications across scientific disciplines.
In microbiome research, the choice of sequencing methodology is foundational, dictating the depth and breadth of biological insight one can attain. While 16S rRNA gene sequencing (16S) has been a workhorse for microbial community profiling, shotgun metagenomic sequencing (shotgun) provides a comprehensive view of all genetic material in a sample, enabling unparalleled functional analysis [10]. This Application Note delineates the technical and practical distinctions between these two principal methods, with a specific emphasis on their utility for inferring and understanding microbial function. The content is framed within the context of advanced research aimed at microbiome functional analysis, providing drug development professionals and scientists with the protocols and data comparisons necessary to inform their experimental design.
Fundamental Technical Differences
The core distinction between these methods lies in their scope and underlying approach. 16S sequencing is a form of amplicon sequencing that relies on the polymerase chain reaction (PCR) to amplify a single, highly conserved gene—the 16S ribosomal RNA gene—which serves as a phylogenetic marker for identifying and quantifying bacteria and archaea [11] [10]. In contrast, shotgun metagenomics is an untargeted approach that involves fragmenting all the DNA in a sample into millions of small pieces, sequencing them, and then reconstructing the genomic content bioinformatically [2] [1]. This fundamental difference is illustrated below.
Quantitative Comparative Analysis
Performance Metrics and Capabilities
A direct comparison of performance metrics is critical for selecting the appropriate methodology. The table below synthesizes key comparative data from recent studies.
Table 1: Comparative performance of 16S rRNA and shotgun metagenomic sequencing
| Feature | 16S rRNA Sequencing | Shotgun Metagenomic Sequencing | Experimental Support |
|---|---|---|---|
| Taxonomic Resolution | Genus-level (species-level possible but with high false positives) [12] | Species and strain-level resolution [13] [12] | Jovel et al., 2016: Shotgun provided improved genus- and species-level classification [10] |
| Taxonomic Coverage | Bacteria and Archaea only [11] [13] | Multi-kingdom: Bacteria, Archaea, Fungi, Viruses, Protists [2] [13] | |
| Functional Profiling | Indirect prediction (e.g., PICRUSt); does not capture true functional diversity [13] [12] | Direct characterization of functional genes and pathways [2] [13] | |
| Sensitivity (Genera Detection) | Detects only part of the microbial community [14] [5] | Higher power to identify less abundant taxa with sufficient reads [14] | Tettamanti et al., 2024: 16S showed sparser data and lower alpha diversity compared to shotgun [5] |
| Quantitative Concordance | Abundance correlated with shotgun for shared taxa, but gives greater weight to dominant bacteria [5] | More comprehensive and symmetrical relative species abundance distribution [14] | Raimondi et al., 2021: Shotgun RSA distributions were more symmetrical; 16S was patchy and skewed [14] |
| Differential Abundance Power | Lower; identified 108 significant genera (caeca vs. crop) [14] | Higher; identified 256 significant genera (caeca vs. crop) [14] | Raimondi et al., 2021: Shotgun found 152 changes missed by 16S; 16S found 4 changes missed by shotgun [14] |
Practical Considerations for Study Design
Beyond performance, several practical factors influence method selection, particularly in a drug development context.
Table 2: Practical considerations for selecting a sequencing method
| Factor | 16S rRNA Sequencing | Shotgun Metagenomic Sequencing |
|---|---|---|
| Cost per Sample | ~$50 USD [13] | Starting at ~$150 USD; shallow shotgun can approach 16S cost for stool samples [13] |
| Bioinformatics | Beginner to intermediate expertise; well-established pipelines (QIIME, MOTHUR) [11] [13] | Intermediate to advanced expertise; more complex pipelines (MetaPhlAn, HUMAnN) [2] [13] |
| Ideal Sample Type | All types, especially low microbial biomass samples (e.g., skin swabs, tissue) [13] [12] | Samples with high microbial biomass (e.g., stool); host DNA removal may be needed for other types [13] [12] |
| Host DNA Interference | Low (PCR step enriches for 16S gene) [12] | High (sequences all DNA); can be mitigated by sequencing depth or host depletion [13] [12] |
| Minimum DNA Input | Low (can be <1 ng due to PCR amplification) [12] | Higher (typically >1 ng/μL); techniques for low biomass exist [12] |
| Reference Databases | Well-curated (SILVA, Greengenes, RDP) [13] [10] | Larger but less curated (RefSeq, GenBank); dependent on genome quality [13] [10] |
Detailed Experimental Protocols
Protocol for 16S rRNA Gene Sequencing
Principle: This protocol involves the targeted amplification and sequencing of hypervariable regions of the bacterial 16S rRNA gene to determine taxonomic composition [11] [15].
Table 3: Research reagents for 16S rRNA sequencing
| Reagent / Kit | Function |
|---|---|
| DNeasy PowerLyzer PowerSoil Kit (Qiagen) or equivalent | DNA extraction from complex samples, removing inhibitors [5] |
| Primers targeting hypervariable regions (e.g., V3-V4, V4) | PCR amplification of specific 16S rRNA gene regions [11] [10] |
| High-Fidelity DNA Polymerase | Accurate amplification of the target region with low error rate [11] |
| Magnetic Beads for Size Selection | Cleanup of PCR amplicons to remove primers and impurities [11] |
| SILVA, Greengenes, or RDP Database | Reference databases for taxonomic classification of sequences [10] [5] |
Procedure:
- Sample Collection and DNA Extraction:
- Collect sample (e.g., stool, swab) using sterile techniques and freeze immediately at -20°C or -80°C to preserve microbial integrity [11].
- Extract genomic DNA using a specialized kit (e.g., DNeasy PowerLyzer Powersoil kit). The choice of extraction kit significantly impacts the microbial profile observed and must be consistent across a study [5].
Library Preparation:
- Amplification: Perform PCR using primers that flank the chosen hypervariable region(s) (e.g., V3-V4) of the 16S rRNA gene [11] [5].
- Indexing: Incorporate unique molecular barcodes (indices) into each sample's amplicons during a second, limited-cycle PCR to enable sample multiplexing [11] [13].
- Clean-up: Purify the final amplicon library using magnetic beads to remove contaminants and select for the correct fragment size [11].
Sequencing:
- Quantify the purified libraries, pool them in equimolar ratios, and sequence on an Illumina MiSeq or similar platform, typically generating 2x250 bp or 2x300 bp paired-end reads [11].
Bioinformatic Analysis:
- Quality Control: Trim primers and filter reads based on quality and length using tools like DADA2 [5].
- Inference: Generate Amplicon Sequence Variants (ASVs) or Operational Taxonomic Units (OTUs) to represent unique biological sequences [14] [5].
- Taxonomy Assignment: Classify ASVs/OTUs by aligning them to a 16S-specific reference database (e.g., SILVA) [10] [5].
- Functional Prediction (Indirect): Use tools like PICRUSt to predict functional potential based on the identified taxonomy, acknowledging this is an inference and not a direct measurement [13].
Protocol for Shotgun Metagenomic Sequencing
Principle: This protocol involves the untargeted sequencing of all DNA fragments in a sample, allowing for simultaneous taxonomic profiling at high resolution and direct assessment of functional potential [2] [1].
Table 4: Research reagents for shotgun metagenomic sequencing
| Reagent / Kit | Function |
|---|---|
| NucleoSpin Soil Kit (Macherey-Nagel) or equivalent | Comprehensive DNA extraction for diverse microbial communities [5] |
| Tagmentation Enzyme / Fragmentation Kit | Random fragmentation of genomic DNA into short inserts [13] |
| Indexing Adapters (Illumina) | Ligation of unique barcodes for sample multiplexing and sequencing primers |
| Bioinformatics Pipelines: MetaPhlAn, Kraken2, HUMAnN2 | Tools for taxonomic profiling and functional analysis from raw reads [2] [13] |
Procedure:
- Sample Collection and DNA Extraction:
- Follow stringent collection and preservation protocols as for 16S. Extract total genomic DNA using a robust kit (e.g., NucleoSpin Soil Kit) designed to lyse a wide range of microbes [5]. For samples with high host DNA (e.g., tissue), consider implementing a host DNA depletion step.
Library Preparation:
- Fragmentation: Fragment the purified DNA mechanically or enzymatically (e.g., via tagmentation) to a desired size (e.g., 300-500 bp) [13].
- Adapter Ligation: Ligate sequencing adapters containing unique dual indices (UDIs) to the fragmented DNA [2] [10].
- Optional PCR: Perform a limited-cycle PCR to amplify the library if input DNA is low.
- Clean-up and Quality Control: Purify the library and validate its size distribution and concentration using an instrument such as a Bioanalyzer.
Sequencing:
- Pool libraries and sequence on a high-throughput platform (e.g., Illumina NovaSeq) to achieve the desired depth. For functional analysis, deeper sequencing (e.g., 10-20 million reads per sample) is often required compared to shallow sequencing for taxonomy alone [1].
Bioinformatic Analysis:
- Pre-processing: Remove low-quality reads and adapter sequences. For clinical samples, filter out reads aligning to the host genome (e.g., GRCh38) using Bowtie2 [5].
- Taxonomic Profiling: Assign reads to taxonomic units using marker-based (e.g., MetaPhlAn) or alignment-based (e.g., Kraken2) tools against genomic databases like RefSeq [2] [10].
- Functional Profiling: Align reads to functional databases (e.g., KEGG, eggNOG) using tools like HUMAnN2 to quantify the abundance of genes and metabolic pathways directly from the metagenomic data [2] [13].
Application in Microbial Functional Analysis
The capacity of shotgun sequencing to directly profile genes confers a decisive advantage for investigating the functional potential of microbiomes, a critical aspect in therapeutic development. Shotgun data can identify antibiotic resistance genes, virulence factors, and biosynthetic pathways for metabolite production [2] [13]. Evidence from large human studies suggests that functional metagenomic data may provide more power for identifying differences between 'healthy' and 'diseased' microbiomes than taxonomic data alone [13] [10].
For instance, in colorectal cancer (CRC) research, shotgun sequencing has been instrumental in defining microbial signatures not only by taxonomy (e.g., Fusobacterium, Parvimonas micra) but also by the collective genetic capabilities of the dysbiotic community [5]. This direct functional insight is unattainable with 16S sequencing, where function must be predicted indirectly, often missing strain-specific functions and novel genes [13].
Shotgun and 16S sequencing provide two distinct lenses for examining microbial communities. 16S rRNA sequencing remains a powerful, cost-effective tool for large-scale, targeted surveys of bacterial and archaeal composition, particularly in studies where budget and computational resources are primary constraints, or for sample types with high host contamination.
However, for research focused on microbiome functional analysis, shotgun metagenomic sequencing is unequivocally the superior choice. Its ability to provide high-resolution taxonomic classification across all domains of life and, most importantly, to directly interrogate the functional gene content of a microbiome, delivers a comprehensive view of the community's genetic potential. As the field of microbiome research increasingly shifts toward understanding function and mechanism in drug development, shotgun metagenomics, including the cost-effective shallow shotgun approach for suitable sample types, is becoming the indispensable standard.
The vast majority of the microbial world has historically been inaccessible to scientific investigation due to one significant challenge: the inability to cultivate these organisms in laboratory settings. This phenomenon, first recognized by Robert Koch in the 19th century when he observed that only a limited number of microorganisms from samples grew on potato substrates, continues to impede research today [16]. Environmental microbiologists estimate that less than 2% of environmental bacteria can be cultured using standard laboratory techniques, while approximately 50% of oral microorganisms resist cultivation, with similar or higher figures anticipated for other body sites like the colon [16]. The significance of this challenge is profound; we remain largely ignorant of bacterial life on Earth, potentially missing novel pathogens, beneficial organisms, and unique metabolic pathways that could revolutionize fields from medicine to biotechnology.
The term "unculturable" does not imply that these microorganisms cannot ever be cultured, but rather that their specific growth requirements are unknown or cannot be replicated with current standard laboratory methodologies [17]. Many exist in a state known as "viable but non-culturable" (VBNC), where they maintain low metabolic activity but refuse to divide on conventional culture media [18]. This VBNC state represents a survival strategy for numerous bacterial species when faced with unfavorable growth conditions, including inappropriate temperature, pH, nutrient limitation, or antibiotic stress [18]. Understanding and overcoming these cultivation limitations is essential for comprehensive microbiome research, particularly in the context of shotgun metagenomic sequencing and functional analysis that aims to characterize complete microbial communities and their interactions with hosts.
Understanding Unculturable Microorganisms
Defining the Unculturable Microbiome
Unculturable microorganisms are those that have not yet been successfully grown and maintained in isolation under controlled laboratory conditions, despite their demonstrable presence and activity in natural environments [19] [18]. Their existence is primarily inferred through culture-independent molecular techniques that detect their genetic material or gene expression directly from environmental samples. This "microbial dark matter" represents a significant knowledge gap in microbiology, as the vast majority of microbial diversity on Earth—estimated to be over 99% of species—remains uncultivated [20] [18]. This limitation profoundly impacts microbiome research, as we cannot fully understand microbial community structure, function, or host-microbe interactions without accessing these hidden members.
Reasons for Uncultivability
Multiple interconnected factors contribute to the challenge of cultivating environmental microorganisms in laboratory settings:
Complex nutritional requirements: Many unculturable microorganisms have highly specific nutrient needs that are not replicated in standard synthetic media. Some have acquired mutations in essential synthetic pathways through evolution and depend on metabolic byproducts from other community members [16]. For instance, Bacteroides forsythus, associated with periodontitis, has an absolute requirement for N-acetyl muramic acid, a peptidoglycan component it cannot synthesize independently [16].
Disrupted bacterial communication networks: Microbes in natural environments exist in complex communication networks mediated by bacterial cytokines and signaling molecules. The separation of bacteria on solid media disrupts these networks, potentially explaining why some organisms fail to grow in isolation [16]. Resuscitation-promoting factors (Rpf) discovered in Micrococcus luteus stimulate growth of other Gram-positive bacteria at picomolar concentrations, demonstrating the importance of such signaling [16].
Inability to simulate native environments: Laboratories cannot fully replicate the complex, dynamic conditions of natural environments, including subtle physicochemical gradients, microenvironments, and community interactions that are essential for some microorganisms [18]. This is particularly challenging for extremophiles adapted to specialized niches with unique combinations of temperature, pressure, salinity, or other factors.
Microbial interdependence: Many microorganisms exist in obligate symbiotic relationships where they depend on other species for essential nutrients, growth factors, or waste removal [16] [17] [18]. The "Black Queen Hypothesis" suggests some microbes shed genes for essential functions, relying instead on other community members to provide these public goods [19].
Low abundance and slow growth rates: In many environments, some microbial species exist at low abundance or have extremely slow growth rates, making them difficult to detect and isolate before faster-growing species dominate the culture [18].
Table 1: Primary Factors Contributing to Microbial Uncultivability
| Factor Category | Specific Challenges | Examples |
|---|---|---|
| Nutritional Requirements | Specific nutrient needs, auxotrophy, unknown growth factors | Bacteroides forsythus requires N-acetyl muramic acid [16] |
| Environmental Conditions | Unable to replicate native physicochemical parameters, oxygen sensitivity | Difficulties culturing anaerobes without proper redox conditions [18] |
| Microbial Interactions | Disrupted signaling, dependency on other species | Catellibacterium nectariphilum requires helper organisms [17] |
| Biological State | Viable but non-culturable (VBNC) state, dormancy | Over 100 species can enter VBNC state [18] |
Shotgun Metagenomics: A Culture-Independent Approach
Principles of Shotgun Metagenomic Sequencing
Shotgun metagenomic sequencing represents a transformative approach for studying unculturable microorganisms by bypassing the need for cultivation entirely. This next-generation sequencing method involves extracting DNA directly from environmental or clinical samples, fragmenting it into small pieces, and sequencing all DNA fragments in parallel [1] [7]. The resulting sequences are then computationally analyzed to reconstruct taxonomic profiles and functional potential without requiring prior knowledge of the organisms present [7]. This approach stands in contrast to targeted methods like 16S rRNA gene sequencing, which only amplifies and sequences specific phylogenetic marker genes, thereby limiting insights into the overall functional capacity of microbial communities [7] [14].
The power of shotgun metagenomics lies in its ability to provide comprehensive sampling of all genes from all organisms in a complex sample [1]. This enables researchers to evaluate bacterial diversity and detect microbial abundance while simultaneously accessing genetic information about functional capabilities, including metabolic pathways, virulence factors, and antibiotic resistance genes [1] [21]. For unculturable microorganisms, this method provides a window into their genetic makeup and potential ecological roles that would otherwise remain inaccessible through culture-dependent approaches.
Comparison with 16S rRNA Sequencing
While 16S rRNA sequencing has been the workhorse of microbial ecology for decades, shotgun metagenomics offers several distinct advantages for studying unculturable microorganisms, particularly in the context of functional analysis:
Table 2: Comparison of 16S rRNA Gene Sequencing vs. Shotgun Metagenomic Sequencing
| Parameter | 16S rRNA Gene Sequencing | Shotgun Metagenomics |
|---|---|---|
| Genetic Target | Amplifies only 16S rRNA hypervariable regions [14] | Sequences all genomic DNA fragments [1] [14] |
| Taxonomic Resolution | Typically genus-level, sometimes species-level [14] | Species to strain-level differentiation possible [21] |
| Functional Insights | Limited to inference from phylogenetic relationships [7] | Direct identification of functional genes and pathways [1] [7] |
| Coverage of Diversity | Primarily bacteria and archaea with universal primers [7] | All domains (bacteria, archaea, viruses, eukaryotes) [21] |
| Detection Sensitivity | May miss low-abundance taxa [14] | Better detection of rare community members with sufficient sequencing depth [14] |
| Quantitative Accuracy | Subject to PCR amplification biases [7] | More directly quantitative, though with own biases [14] |
Research comparing these methodologies directly demonstrates that shotgun sequencing detects a substantially greater proportion of microbial diversity. One study found that shotgun sequencing identified 256 statistically significant differences in genera abundance between gut compartments, while 16S sequencing detected only 108 differences [14]. Additionally, the less abundant genera detected exclusively by shotgun sequencing proved biologically meaningful in discriminating between experimental conditions [14].
Diagram 1: Shotgun Metagenomic Sequencing Workflow
Experimental Protocols for Studying Unculturable Microbes
Sample Collection and DNA Extraction
Proper sample collection and processing are critical for accurate metagenomic analysis, as biases introduced at these stages can profoundly affect downstream results:
Sample Collection: Collect samples using standardized methods appropriate for the environment being studied (e.g., sterile swabs for body sites, core samplers for soil, filtration for water) [21]. Immediately preserve samples using stabilization solutions like DNA/RNA Shield to prevent microbial community shifts and nucleic acid degradation [21]. For human gut microbiota studies, sample multiple gastrointestinal compartments when possible, as community composition varies significantly between locations [14].
DNA Extraction: Utilize mechanical and enzymatic lysis methods capable of breaking diverse cell walls (Gram-positive, Gram-negative, fungal) [21]. The ZymoBIOMICS DNA Mini Kit and similar systems provide standardized protocols for efficient DNA extraction from complex samples. Include appropriate controls such as extraction blanks to detect contamination and standardized microbial communities (ZymoBIOMICS Microbial Community Standard) to assess technical variability and efficiency [21]. Quantify DNA using fluorescence-based methods (e.g., Qubit) rather than UV absorbance, as the latter is less accurate for complex microbial mixtures.
Library Preparation and Sequencing
Library preparation converts extracted DNA into sequencing-ready fragments:
DNA Fragmentation: Use controlled enzymatic or mechanical fragmentation to generate appropriately sized DNA fragments (typically 200-500bp for Illumina platforms) [20]. The Illumina DNA Prep kit provides a standardized approach for this process.
Adapter Ligation: Attach platform-specific sequencing adapters with unique dual indexes (UDIs) to enable sample multiplexing and prevent index hopping issues [21]. UDIs are essential for accurate sample identification in pooled sequencing runs.
Sequencing Parameters: For complex microbial communities, aim for sufficient sequencing depth to detect rare community members. While requirements vary by community complexity, studies suggest a minimum of 500,000 reads per sample for meaningful analysis, with 2-5 million reads providing better coverage for diverse communities [20] [14]. Paired-end sequencing (2×150 bp) on Illumina platforms (NovaSeq, NextSeq) provides sufficient read length and quality for most metagenomic applications [21].
Bioinformatic Analysis Pipeline
The computational analysis of metagenomic data involves multiple steps to transform raw sequences into biological insights:
Quality Control and Preprocessing: Process raw sequencing reads using tools like FastQC for quality assessment and Trimmomatic or Cutadapt for adapter removal and quality filtering. Remove host-derived sequences if working with host-associated samples using tools like DeconSeq or Bowtie2 alignment to host genomes [7].
Assembly and Binning: For high-complexity samples, assemble quality-filtered reads into contigs using metaSPAdes or MEGAHIT [7] [20]. The success of assembly varies dramatically with community complexity—while simple communities like acid mine drainage biofilms showed 85% of reads assembling into contigs, highly diverse soil communities may see less than 1% assembly [20]. Group contigs into genome bins based on sequence composition (GC content, k-mer frequencies) and abundance profiles across samples [7] [20].
Taxonomic Profiling: Classify sequences using reference-based methods against comprehensive databases. K-mer based approaches like those implemented in SourMash provide strain-level taxonomic resolution when using databases such as GTDB (Genome Taxonomy Database) or NCBI that contain over 77,000 reference strains across all kingdoms [21].
Functional Annotation: Identify protein-coding genes in assembled contigs or directly from reads using Prodigal or FragGeneScan. Annotate predicted genes against databases like UniRef (gene families), KEGG (pathways), and MetaCyc (metabolic pathways) to determine functional potential [7] [21]. Specialized databases like CARD for antibiotic resistance genes and VFDB for virulence factors enable detection of specific functions relevant to human health [21].
Table 3: Key Bioinformatics Tools for Metagenomic Analysis
| Analysis Step | Recommended Tools | Primary Function |
|---|---|---|
| Quality Control | FastQC, Trimmomatic, Cutadapt | Assess read quality, remove adapters, filter poor sequences |
| Host Decontamination | DeconSeq, Bowtie2 | Identify and remove host-derived sequences |
| Assembly | metaSPAdes, MEGAHIT | Reconstruct longer contigs from short reads |
| Binning | MetaBAT2, MaxBin2 | Group contigs into putative genomes |
| Taxonomic Classification | SourMash, Kraken2, MetaPhlAn | Assign taxonomy to sequences |
| Functional Annotation | Prodigal, HUMAnN3, DIAMOND | Identify genes and metabolic pathways |
| Visualization & Statistics | Phyloseq, LEfSe, STAMP | Compare communities, identify differentially abundant features |
Applications in Drug Development and Microbiome Research
Pharmacomicrobiomics: Drug-Microbiome Interactions
The study of how genetic and phenotypic diversity in the microbiome affects therapeutic outcomes—pharmacomicrobiomics—represents a promising application of metagenomics in drug development [22]. The human microbiome encodes a vast repertoire of enzymatic activities that can directly modify drug compounds, affecting their efficacy, toxicity, and pharmacokinetics [22]. At least 50 drugs are known to be metabolized by bacteria, though in most cases the specific microbial species and genetic determinants remain unidentified [22]. Notable examples include the activation of the antiviral sorivudine by gut bacterial metabolism, which can lead to toxic interactions with fluoropyrimidine chemotherapy, and the inactivation of the cardiac drug digoxin by specific gut bacterial strains [22].
Shotgun metagenomics enables comprehensive profiling of these drug-microbiome interactions through several approaches:
Culture Collection Screens: High-throughput coincubation of representative human gut bacterial strains with drug compounds, followed by liquid chromatography-mass spectrometry to monitor drug transformation [22]. This approach directly identifies metabolically active strains but provides limited sampling of microbial diversity.
Ex Vivo Fecal Incubations: Incubation of drugs with complex stool samples to capture community-level metabolism, though interstrain antagonism may mask some metabolic activities [22].
Functional Metagenomics: Cloning of microbial DNA from unculturable organisms into suitable hosts (typically E. coli) followed by screening for drug-metabolizing activities [22]. This approach can directly link metabolic functions to genetic elements without requiring cultivation of the source organism.
Metagenome-Wide Association Studies: Correlation of microbial genes and pathways with drug response variability in human populations, potentially identifying microbial biomarkers for treatment personalization [22].
Mining Unculturable Microbes for Novel Therapeutics
Unculturable microorganisms represent an extensive untapped resource for novel therapeutic compounds, including antibiotics, immunosuppressants, and anti-cancer agents. The biosynthetic gene clusters (BGCs) encoding these compounds can be identified directly from metagenomic data, even when the producing organisms cannot be cultured [20]. Studies of diverse environments have revealed that uncultured microorganisms harbor a tremendous diversity of BGCs, many with novel architectures suggesting previously unknown chemical entities.
Functional metagenomics enables the discovery of these compounds by expressing DNA from uncultured microorganisms in heterologous hosts. This approach involves extracting environmental DNA, cloning large fragments into bacterial artificial chromosomes or other vectors, transforming suitable host strains, and screening for bioactivities of interest [20]. Successes include the discovery of novel antibiotics such as turbomycins and terragines from soil metagenomes, demonstrating the potential of this approach for drug discovery [20].
Research Reagent Solutions for Metagenomic Studies
Table 4: Essential Research Reagents and Tools for Metagenomic Studies
| Reagent/Tool | Function | Examples/Specifications |
|---|---|---|
| Sample Preservation Solutions | Stabilize microbial community composition at collection, prevent nucleic acid degradation | DNA/RNA Shield (Zymo Research) [21] |
| DNA Extraction Kits | Efficient lysis of diverse microorganisms, minimal bias | ZymoBIOMICS DNA Mini Kit [21] |
| Library Preparation Kits | Fragment DNA, add sequencing adapters, index samples | Illumina DNA Prep with Unique Dual Indexes [21] |
| Metagenomic Standards | Control for technical variability, assess process efficiency | ZymoBIOMICS Microbial Community Standard [21] |
| Reference Databases | Taxonomic classification and functional annotation | GTDB, NCBI, UniRef, KEGG, MetaCyc [21] |
| Bioinformatics Pipelines | Integrated analysis of metagenomic data | DRAGEN Metagenomics, HUMAnN3, SourMash [1] [21] |
Shotgun metagenomic sequencing has revolutionized our ability to study unculturable microorganisms, providing unprecedented access to the genetic diversity and functional potential of previously inaccessible microbial dark matter. By combining sophisticated molecular techniques with advanced computational analysis, researchers can now characterize microbial community membership, reconstruct metabolic pathways, and identify novel genes of biomedical interest—all without requiring laboratory cultivation of the source organisms.
The implications for drug development and personalized medicine are profound. As we deepen our understanding of how unculturable members of the human microbiome influence drug metabolism, disease susceptibility, and treatment outcomes, we move closer to truly personalized therapeutic approaches that account for both human and microbial genetic variation. The continued development of cultivation techniques, sequencing technologies, and bioinformatic tools will further enhance our ability to mine this untapped resource, potentially yielding novel therapeutic compounds and transformative insights into host-microbe interactions that shape human health and disease.
Shotgun metagenomic sequencing represents a transformative approach in microbial ecology, enabling comprehensive analysis of genetic material directly from environmental, clinical, or industrial samples [23]. Unlike targeted amplicon sequencing (e.g., 16S rRNA gene sequencing), this technique sequences all genomic DNA present in a sample, providing unprecedented access to the taxonomic composition and functional potential of complex microbial communities [2]. The term "shotgun" derives from the process of fragmenting DNA into numerous small pieces that are sequenced and subsequently reassembled computationally [2]. This culture-independent method has revolutionized our understanding of microbial ecosystems across diverse fields including human health, environmental microbiology, and biotechnology [23] [2].
The analytical pipeline for shotgun metagenomic data constitutes a critical framework for transforming raw genetic fragments into biologically meaningful insights. This multi-step process involves quality control, taxonomic profiling, functional annotation, and advanced analyses such as strain-level characterization and metabolic pathway reconstruction [24] [23]. The selection of appropriate tools and parameters at each stage significantly impacts the accuracy and biological relevance of the final results, making a thorough understanding of the entire workflow essential for researchers embarking on microbiome studies [25] [26].
Experimental Design and Sample Preparation
Sample Collection Considerations
Rigorous sample collection protocols are fundamental to obtaining reliable metagenomic data. Three critical factors must be considered during sample acquisition: sterility, temperature, and timing [2]. Sample containers must be sterile to prevent contamination from exogenous microbes. Temperature control is essential for preserving microbial integrity; samples should be frozen immediately at -20°C or -80°C, or snap-frozen in liquid nitrogen. Freeze-thaw cycles adversely affect microbiome consistency, making aliquoting advisable before freezing. When immediate freezing is impossible, temporary storage at 4°C or preservation buffers can maintain sample integrity for hours to days before processing [2].
Sample selection must account for substantial temporal and spatial variations in microbiomes. Consistent sampling protocols across a study population ensure comparability between samples. In clinical contexts such as the study of gut microbiota during acute pancreatitis recovery, rectal swabs collected following standardized procedures (cleaning with soap, water, and 70% alcohol, followed by sterile swab insertion to 4-5 cm depth) provide representative samples when direct fecal collection is impractical [27].
DNA Extraction and Library Preparation
DNA extraction typically employs commercial kits using combined chemical and mechanical methods to lyse cells, precipitate DNA, and purify nucleic acids [2]. The choice of extraction kit significantly influences the observed microbial community structure, affecting inter-study comparisons. Difficult-to-lyse structures (e.g., spores) may require additional enzymatic or heat treatments. For shotgun metagenomics, library preparation involves fragmenting DNA (mechanically or enzymatically), ligating molecular barcodes (index adapters) for sample multiplexing, and cleanup to ensure appropriate fragment size and purity [2].
Computational Analysis Pipeline
Quality Control and Host DNA Removal
Quality control represents the critical first step in computational analysis, addressing sequencing errors that can overestimate microbial diversity and cause erroneous taxonomic assignments [23]. Tools like fastp [27], Trimmomatic [25], and FastQC [25] perform adapter trimming, quality filtering, and read length selection. The fastp tool (v0.23.0) efficiently removes sequencing adapters, eliminates low-quality reads (average quality score <20), and discards short sequences (<50 bp after trimming) [27].
Host DNA removal is particularly important for samples with high host contamination (e.g., clinical specimens from skin or tissues). KneadData [25], BWA [27], or Bowtie2 [26] align reads against host reference genomes (e.g., human) to identify and remove contaminating sequences. In the acute pancreatitis study, BWA (v0.7.17) mapped reads to the human genome, effectively depleting host-derived sequences [27].
Table 1: Essential Tools for Metagenomic Data Preprocessing
| Tool | Function | Key Parameters | Citation |
|---|---|---|---|
| fastp | Quality control and adapter trimming | Quality threshold (Q20), min length 50 bp | [27] |
| FastQC | Quality assessment | Generates interactive QC reports | [25] |
| KneadData | Host read removal | Reference database of host genome | [25] |
| Bowtie2 | Host read alignment and removal | Sensitive local alignment | [26] |
| BWA | Host read removal | Efficient mapping to reference | [27] |
Taxonomic Profiling
Taxonomic classification assigns microbial identities to sequencing reads, enabling community composition analysis. Two primary approaches exist: read-based classification using marker genes or whole genomes, and assembly-based approaches that reconstruct longer sequences before classification [26].
MetaPhlAn4 utilizes unique clade-specific marker genes for efficient taxonomic profiling [25]. Kraken2 employs k-mer-based classification against comprehensive genomic databases [25]. Meteor2 represents a recent advancement that leverages environment-specific microbial gene catalogs for enhanced sensitivity, particularly for low-abundance species [24]. Benchmark tests demonstrate that Meteor2 improves species detection sensitivity by at least 45% compared to MetaPhlAn4 in both human and mouse gut microbiota simulations [24].
Table 2: Taxonomic Profiling Tools and Performance Characteristics
| Tool | Method | Database | Sensitivity Improvement | Citation |
|---|---|---|---|---|
| MetaPhlAn4 | Clade-specific marker genes | ChocoPhlAn | Baseline | [25] [24] |
| Kraken2 | k-mer matching | Custom genomic database | Not specified | [25] |
| Meteor2 | Environment-specific gene catalogs | Custom MSP-based | 45% vs MetaPhlAn4 | [24] |
| Kaiju | Protein-level classification | Reference protein databases | High MCC at species/genus | [26] |
Functional Annotation
Functional annotation identifies metabolic capabilities and biochemical pathways within microbial communities, connecting taxonomic composition to potential ecosystem functions [28]. HUMAnN3 is a widely-used pipeline that maps reads to protein families (UniRef90) and pathway databases (MetaCyc) to quantify functional potential [25]. Meteor2 provides integrated taxonomic, functional, and strain-level profiling (TFSP) using microbial gene catalogs annotated with KEGG Orthology (KO), carbohydrate-active enzymes (CAZymes), and antibiotic resistance genes (ARGs) [24].
In functional abundance estimation, Meteor2 demonstrates 35% improved accuracy compared to HUMAnN3 based on Bray-Curtis dissimilarity metrics [24]. eggNOG-mapper and GO FEAT offer alternative functional annotation approaches, reporting Orthologous Groups identifiers and Gene Ontology (GO) terms, respectively [26].
Metagenome Assembly and Binning
For deeper insights into microbial communities, especially for discovering novel organisms, metagenome assembly reconstructs longer contiguous sequences (contigs) from short reads [23]. MEGAHIT and MetaSPAdes are widely used assemblers that efficiently handle complex metagenomic data [25]. Subsequent binning groups contigs into metagenome-assembled genomes (MAGs) using sequence composition and abundance patterns across samples [23].
MetaWRAP provides a comprehensive binning pipeline that refines and evaluates MAGs [25]. In the acute pancreatitis study, researchers mapped quality-controlled reads to representative genes with 95% identity using SOAPaligner (v2.21) to assess gene abundance [27]. EasyMetagenome supports both assembly-based analysis and binning, incorporating tools like MetaProdigal for gene prediction, CD-HIT for gene clustering, and Salmon for contig coverage estimation [25].
Integrated Analysis Pipelines
All-in-One Pipeline Solutions
Several integrated pipelines streamline the entire metagenomic analysis workflow, providing standardized approaches from raw reads to biological interpretation:
EasyMetagenome offers a user-friendly, flexible pipeline supporting quality control, read-based analysis, assembly-based analysis, and binning [25]. It includes over 150 bioinformatics tools and generates publication-ready visualizations. The pipeline is freely available at https://github.com/YongxinLiu/EasyMetagenome [25].
MEDUSA performs comprehensive metagenomic analysis including preprocessing, assembly, alignment, taxonomic classification, and functional annotation [26]. It supports user-built dictionaries to transfer annotations between different functional identifiers, enhancing flexibility in results interpretation.
MetaLAFFA specializes in annotating functional capacities in shotgun metagenomic data with native compute cluster integration [28]. It processes raw FASTQs through host read filtering, duplicate read removal, quality trimming, and functional annotation using KEGG orthology groups and pathway mappings.
Meteor2 employs environment-specific microbial gene catalogs for unified taxonomic, functional, and strain-level profiling [24]. Its database currently includes 10 ecosystems with 63,494,365 microbial genes clustered into 11,653 metagenomic species pangenomes (MSPs).
Table 3: Integrated Metagenomic Analysis Pipelines
| Pipeline | Key Features | Supported Analyses | Installation | Citation |
|---|---|---|---|---|
| EasyMetagenome | User-friendly, 150+ tools | QC, read-based, assembly-based, binning | GitHub repository | [25] |
| MEDUSA | Flexible functional annotation | Preprocessing, assembly, taxonomy, function | Conda | [26] |
| MetaLAFFA | Cluster integration | Functional annotation, pathway mapping | Conda | [28] |
| Meteor2 | Environment-specific gene catalogs | Taxonomic, functional, strain-level profiling | Open-source | [24] |
Data Visualization and Interpretation
Effective visualization is crucial for interpreting complex metagenomic data. ggplot2 in R provides powerful, customizable graphics for creating publication-quality figures [29]. Alpha-diversity analysis (e.g., Shannon index) using R packages "vegan" and "reshape2" quantifies within-sample microbial diversity [27]. Principal coordinates analysis (PCoA) visualizes beta-diversity patterns, revealing similarities and differences in microbial community structure across samples [27].
Color selection in biological data visualization should follow established guidelines to enhance clarity and accessibility [30]. Key principles include: identifying data nature (nominal, ordinal, interval, ratio), selecting appropriate color spaces (e.g., CIE Lab/Luv for perceptual uniformity), checking color context, assessing color deficiencies, and ensuring web/print compatibility [30].
Table 4: Essential Research Reagent Solutions for Shotgun Metagenomics
| Category | Item | Function | Example/Specification |
|---|---|---|---|
| Sample Collection | Sterile containers | Prevent exogenous contamination | DNA/RNA-free tubes |
| Preservation buffers | Maintain sample integrity before freezing | Commercial stabilization solutions | |
| DNA Extraction | DNA extraction kits | Nucleic acid isolation from complex samples | MP-soil FastDNA Spin Kit for Soil [27] |
| Enzymatic treatments | Enhance lysis of resistant structures | Lysozyme, proteinase K | |
| Library Preparation | Fragmentation enzymes | DNA shearing for library construction | Mechanical or enzymatic fragmentation |
| Index adapters | Sample multiplexing | Unique molecular barcodes | |
| Sequencing | Sequencing platforms | High-throughput DNA sequencing | Illumina HiSeq 4000 [27] |
| Control libraries | Quality assessment of sequencing run | PhiX control library | |
| Computational | Reference databases | Taxonomic and functional classification | KEGG [27], GTDB, UniRef90 [28] |
| Analysis pipelines | Streamlined data processing | EasyMetagenome [25], MEDUSA [26] |
Application Case Study: Gut Microbiota in Acute Pancreatitis Recovery
A recent investigation exemplified the complete shotgun metagenomic workflow in studying gut microbiota dynamics during acute pancreatitis (AP) recovery [27]. Researchers collected rectal swabs from 12 AP patients of varying severity during both acute and recovery phases, with four healthy controls. After DNA extraction using MP-soil FastDNA Spin Kit, paired-end sequencing was performed on an Illumina HiSeq 4000 platform [27].
The analytical pipeline included: (1) quality control with fastp (v0.23.0) including adapter removal, quality filtering (Q20 threshold), and length selection (>50 bp); (2) host DNA removal using BWA alignment to human genome; (3) gene abundance assessment via SOAPaligner mapping to representative genes (95% identity); (4) taxonomic and functional profiling; (5) statistical analysis including α-diversity (Shannon index) and PCoA using R packages "vegan" and "reshape2"; and (6) functional annotation against KEGG database using Diamond [27].
This comprehensive approach revealed that during AP recovery, microbial diversity remained decreased with increasing beneficial bacteria (Bacteroidales) in mild AP but elevated harmful bacteria (Enterococcus) in severe cases. Functional analysis identified signaling pathways showing opposite trends in recovery phases, providing insights into microbial community restructuring during convalescence [27].
The analytical pipeline for shotgun metagenomic sequencing represents a sophisticated framework transforming raw genetic data into biological understanding. From meticulous sample preparation through computational analysis to visual interpretation, each stage requires careful consideration of tools and parameters appropriate for the specific research question. Current pipelines like EasyMetagenome, MEDUSA, and Meteor2 offer increasingly integrated solutions, while functional annotation tools like HUMAnN3 and Meteor2 provide deeper insights into microbial community capabilities.
As sequencing technologies evolve and computational methods advance, the pipeline from genetic fragments to biological meaning will continue to refine our understanding of complex microbial ecosystems across diverse environments. The integration of long-read sequencing, improved reference databases, and machine learning approaches promises even more sophisticated metagenomic analyses in the future, further illuminating the invisible microbial world that sustains planetary health and human wellbeing.
Shotgun metagenomic sequencing has revolutionized microbiome research by enabling comprehensive analysis of all genetic material within a sample, moving beyond the limitations of targeted 16S rRNA gene sequencing [31] [1]. This approach allows researchers to characterize thousands of organisms in parallel, providing unprecedented insights into community biodiversity and functional potential [1]. Unlike 16S sequencing, which targets a single phylogenetic marker gene, shotgun sequencing captures the entire genomic content, enabling species-level identification and functional profiling of microbial communities [31] [32]. The analysis of this complex data relies on several fundamental bioinformatics processes, including the reconstruction of Metagenome-Assembled Genomes (MAGs), binning, functional annotation, and taxonomic classification, which together form the foundation for interpreting microbial community structure and function.
Terminology Definitions and Applications
Metagenome-Assembled Genomes (MAGs)
Metagenome-Assembled Genomes (MAGs) represent individual microbial genomes reconstructed from complex metagenomic sequencing data through computational methods [31]. This process bypasses the need for laboratory cultivation, which is particularly valuable since an estimated 1-2% of bacteria can be cultured using standard methods [32]. MAGs are binned into similar bacterial species or strains based on sequence similarity characteristics and coverage, providing insights into previously unculturable and unknown microorganisms [31]. The concept of MAGs has been further refined with the introduction of species-level genome bins (SGBs), which are categorized as known SGBs (kSGBs) for those present in reference databases and unknown SGBs (uSGBs) for newly-assembled genomes not previously cataloged [31]. MAGs enable strain-level analysis and the identification of specific microorganisms that play key roles in various biological processes, from human health to environmental ecosystems [33].
Binning
Binning is the computational process of grouping assembled DNA sequences (contigs) into biologically meaningful clusters that potentially represent individual microbial genomes or population-level genomes [31] [34]. This process typically relies on sequence composition (e.g., GC content, k-mer frequencies) and abundance patterns (coverage depth across multiple samples) to distinguish between different taxonomic origins [31]. Binning is essential for MAG reconstruction because metagenomic assembly produces contigs from all organisms in a community simultaneously without inherent taxonomic separation [34]. Advanced binning approaches now also incorporate taxonomic signals from longer sequences where available, first assigning contigs to MAGs and then associating reads with these longer sequences for more reliable taxonomic annotation [34]. Proper binning reduces data complexity and minimizes the generation of chimeric sequences where fragments from different genomes are incorrectly merged [32].
Functional Annotation
Functional annotation refers to the process of identifying putative biological functions of genes predicted from MAGs or assembled contigs based on information available in reference databases [35]. This process typically involves predicting genes in the assembled sequences and then comparing the protein sequences against reference databases to assign functional categories [35]. Current tools like DRAM perform comprehensive functional annotation using multiple databases including Pfam, KEGG, UniProt, CAZY, and MEROPS [35]. Functional annotation enables researchers to decipher the metabolic capabilities of microbial communities, identifying functions such as KEGG orthology groups, carbohydrate-active enzymes (CAZymes), and antibiotic resistance genes (ARGs) [24]. These annotations form the basis for downstream analyses including functional gene enrichment studies and the distillation of genome-inferred functional traits [35].
Taxonomic Classification
Taxonomic classification in metagenomics involves assigning taxonomic labels (e.g., phylum, genus, species) to sequencing reads, contigs, or MAGs based on their similarity to reference sequences [31] [34]. This process can utilize different approaches including marker gene methods (e.g., MetaPhlAn which uses species-specific marker genes), k-mer based classification (e.g., Kraken2), and homology searches against comprehensive protein databases (e.g., DIAMOND) [31] [34]. Unlike 16S rRNA sequencing, which provides phylogenetic information based on a single gene, shotgun metagenomic taxonomic classification leverages information from across the entire genome, providing significantly improved species-level resolution [31] [36]. Accurate taxonomic classification is essential for understanding microbial community composition and dynamics, with applications ranging from clinical diagnostics to environmental monitoring [36].
Quantitative Performance of Metagenomic Tools
Table 1: Performance comparison of taxonomic profiling tools based on benchmarking studies
| Tool | Classification Approach | Key Strengths | Reported Performance Metrics |
|---|---|---|---|
| MetaPhlAn4 | Marker gene + MAG-based [31] | Well-established, commonly used [31] | Best overall performance in accuracy metrics [31] |
| Meteor2 | Microbial gene catalogues [24] | Excellent for low-abundance species [24] | 45% higher species detection sensitivity in shallow-sequenced data; 35% improved abundance estimation accuracy [24] |
| JAMS | k-mer based (Kraken2) + assembly [31] | High sensitivity [31] | Among highest sensitivity metrics [31] |
| WGSA2 | k-mer based (Kraken2) [31] | High sensitivity [31] | Among highest sensitivity metrics [31] |
| RAT | Integrated MAG/contig/read signals [34] | High precision and sensitivity [34] | Outperforms other tools in precision and sensitivity [34] |
Table 2: Functional annotation databases and their applications
| Database | Primary Focus | Utility in Metagenomics |
|---|---|---|
| KEGG | Metabolic pathways and ortholog groups [35] [24] | Functional orthologs and pathway analysis [24] |
| CAZy | Carbohydrate-active enzymes [24] | Carbohydrate metabolism and degradation [24] |
| MEROPS | Proteolytic enzymes [35] | Protein degradation and peptide metabolism [35] |
| Pfam | Protein families and domains [35] | Protein family classification and domain analysis [35] |
| Resfinder | Antibiotic resistance genes [24] | Identification of clinically relevant ARGs [24] |
Experimental Protocols
Protocol 1: Taxonomic Profiling Using Integrated MAG and Read Signals
Principle: This protocol leverages the RAT (Read Annotation Tool) pipeline to integrate robust taxonomic signals from MAGs and contigs with direct read annotations for comprehensive taxonomic profiling [34].
Procedure:
- Assembly and Binning: Perform de novo assembly of quality-filtered reads using an assembler such as MEGAHIT or metaSPAdes [34]. Bin resulting contigs into MAGs using binning tools like MetaBAT2 or MaxBin [34].
- Contig and MAG Annotation: Annotate contigs and MAGs with CAT (Contig Annotation Tool) and BAT (Bin Annotation Tool) [34]:
- Predict Open Reading Frames (ORFs) on contigs and MAGs using Prodigal [34].
- Query predicted protein sequences against a reference protein database (NCBI nr or GTDB) using DIAMOND blastp [34].
- Assign taxonomy based on the combined taxonomic signals of individual ORFs, selecting higher-ranking taxa when conflicting signals are present [34].
- Read Mapping and Inheritance: Map individual reads to contigs using BWA-MEM, allowing each read to inherit the taxonomic annotation with the highest reliability (MAG annotation for binned contigs, contig annotation for unbinned contigs) [34].
- Direct Read Annotation: For sequences not mapping to contigs or without CAT annotation, perform direct annotation by querying against the protein database with DIAMOND blastx [34].
- Profile Reconstruction: Generate the final taxonomic profile by combining annotations from all steps, prioritizing the most reliable signals first [34].
Protocol 2: Functional Annotation of MAGs Using DRAM
Principle: This protocol uses the DRAM tool to perform comprehensive functional annotation of MAGs, integrating multiple reference databases to assign biological functions to predicted genes [35].
Procedure:
- Gene Prediction: Identify protein-coding genes in MAGs using ab initio gene prediction tools. Gene prediction is typically performed as part of the DRAM pipeline [35].
- Database Searching: Query predicted protein sequences against multiple functional databases simultaneously:
- Annotation Integration: Consolidate results from all databases, resolving conflicting annotations based on statistical confidence metrics and database priority rules established in the DRAM pipeline [35].
- Functional Profiling: Generate a functional profile for each MAG, categorizing genes into metabolic pathways, functional modules, and specific enzyme classes to enable comparative analysis across samples [35].
Protocol 3: Strain-Level Profiling with Meteor2
Principle: This protocol utilizes Meteor2 for strain-level analysis by tracking single nucleotide variants (SNVs) in signature genes of Metagenomic Species Pan-genomes (MSPs) [24].
Procedure:
- Read Mapping: Map quality-trimmed metagenomic reads against a microbial gene catalogue using bowtie2 with alignment parameters set to 95% identity for default mode or more stringent 98% identity for fast mode [24].
- Gene Quantification: Calculate gene counts using one of three computation modes:
- Taxonomic Profiling: Normalize gene count tables and reduce to generate MSP profiles by averaging the abundance of signature genes within each MSP after normalization [24].
- Strain Tracking: Identify strain-level variation by tracking SNVs in the signature genes of MSPs, enabling high-resolution analysis of microbial community dynamics [24].
Workflow Visualization
Shotgun Metagenomics Workflow
MAG Reconstruction & Annotation
Functional Annotation Process
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Essential research reagents and computational tools for shotgun metagenomics
| Category | Specific Products/Tools | Function and Application |
|---|---|---|
| DNA Extraction Kits | MoBIO DNA Extraction Kit, Qiagen DNA Microbiome Kit, Epicentre Metagenomic DNA Isolation Kit [32] | Isolation of high-quality microbial DNA from complex environmental samples; critical for minimizing bias in microbial diversity assessment [32] |
| Library Preparation | Bioo Scientific NEXTflex PCR-Free, Illumina TruSeq PCR-Free, Kapa Hyper Prep Kit [32] | Preparation of sequencing libraries; PCR-free methods recommended to avoid amplification biases when sufficient DNA is available [32] |
| Taxonomic Profilers | MetaPhlAn4, RAT, JAMS, WGSA2, Meteor2 [31] [24] [34] | Assignment of taxonomic labels to reads, contigs, or MAGs; tools vary in approach (marker gene, k-mer, alignment-based) and performance characteristics [31] |
| Functional Annotation Tools | DRAM, Meteor2, HUMAnN3 [35] [24] | Prediction and categorization of gene functions using reference databases; enables metabolic reconstruction and functional potential assessment [35] |
| Reference Databases | KEGG, CAZy, Pfam, MEROPS, GTDB, NCBI nr [35] [24] [34] | Collections of curated biological information used for taxonomic assignment and functional annotation; database selection influences results interpretation [35] |
| DBCO-PEG5-NHS ester | DBCO-PEG5-NHS ester, MF:C36H43N3O11, MW:693.7 g/mol | Chemical Reagent |
| Decafentin | Decafentin, CAS:15652-38-7, MF:C46H51BrClPSn, MW:868.9 g/mol | Chemical Reagent |
From Sample to Insight: Methodological Workflows and Pharmaceutical Applications
Shotgun metagenomic sequencing is a powerful, high-throughput genomic technology that enables researchers to directly access the genetic content of entire microbial communities from complex samples without the need for cultivation [37]. This approach allows for the comprehensive sampling of all genes in all organisms present, providing unparalleled insights into microbial diversity, community structure, and functional potential [1]. Unlike targeted methods such as 16S rRNA sequencing, shotgun metagenomics captures data from all genomic domains—including bacteria, fungi, viruses, and archaea—while also enabling the reconstruction of metabolic pathways and the discovery of novel genes [37] [38]. This methodological framework is particularly valuable for microbiome functional analysis research, as it facilitates a systems-level understanding of how microbial communities operate and interact within their environments, from the human gut to various ecological niches.
Experimental Design and Sample Collection
The initial phase of any successful shotgun metagenomics study involves careful experimental design and sample collection, as these steps fundamentally influence all subsequent analyses. Sample processing represents the first and most critical step, where the goal is to obtain DNA that is representative of all cells present in the sample while ensuring sufficient quantities of high-quality nucleic acids for library production and sequencing [37].
Table 1: Sample Collection and Processing Considerations for Different Sample Types
| Sample Type | Key Considerations | Recommended Methods |
|---|---|---|
| Stool | Standardized collection; stable DNA | Commercial kits; homogenization |
| Skin (Low-Biomass) | High host contamination; minimal microbial DNA | D-Squame discs; in-house DNA extraction protocols [38] |
| Soil | Coextraction of enzymatic inhibitors (e.g., humic acids) | Physical separation of cells from soil matrix; benchmarked extraction procedures [37] |
| Host-Associated | Overwhelming host DNA | Fractionation or selective lysis to minimize host DNA [37] |
For low-biomass samples like skin swabs, specific collection methods such as D-Squame discs have proven most effective for maximizing DNA yields [38]. When dealing with samples containing inhibitory substances, physical separation of cells from the matrix (e.g., soil) before lysis can reduce coextraction of contaminants that interfere with downstream processing [37]. For host-associated microbiomes, fractionation or selective lysis methods are crucial to minimize host DNA contamination, which could otherwise overwhelm the microbial signal in sequencing data [37].
DNA Extraction and Quality Control
DNA extraction represents a potential source of significant bias in metagenomic studies, as different protocols can vary considerably in their efficiency across diverse microbial taxa. The extraction method must be carefully selected to ensure representative lysis of all cell types present in the community while yielding DNA of sufficient quantity and quality for library preparation.
Multiple studies have demonstrated that direct lysis of cells within the sample matrix versus indirect lysis after cell separation produces quantifiable differences in microbial diversity, DNA yield, and sequence fragment length [37]. For low-biomass samples like skin microbiomes, research indicates that specialized in-house DNA extraction protocols often outperform standardized kits when it comes to maximizing microbial DNA yields [38]. A critical finding from recent work is that Multiple Displacement Amplification (MDA), while capable of amplifying femtograms of DNA to micrograms of product, introduces significant compositional biases and is not recommended for metagenomic applications where accurate representation of community structure is essential [37] [38].
For samples yielding very small amounts of DNA, amplification might be necessary, but researchers should be aware of potential problems including reagent contamination, chimera formation, and sequence bias, all of which can significantly impact subsequent metagenomic community analysis [37]. Quality control assessment of extracted DNA typically involves fluorometric quantification, fragment analysis, and purity assessment via spectrophotometric methods to ensure the material is suitable for library construction.
Library Preparation and Sequencing Technologies
Library preparation converts the extracted DNA into a format compatible with sequencing platforms, with specific protocols varying based on the technology employed. This process typically involves DNA fragmentation, size selection, adapter ligation, and potentially amplification steps. The choice of sequencing technology represents a critical decision point that balances read length, accuracy, throughput, and cost considerations for metagenomic applications.
Table 2: Comparison of Sequencing Technologies for Metagenomics
| Technology | Read Length | Key Features | Best Suited For |
|---|---|---|---|
| Short-Read (Illumina) | 150-300 bp (paired-end) | High accuracy; very high throughput; low cost per Gb [37] [1] | Taxonomic profiling; gene abundance studies; large-scale comparative analyses |
| Long-Read (PacBio HiFi) | Several kilobases | High accuracy long reads; excellent for assembly [33] | Metagenome-assembled genomes (MAGs); resolving complex genomic regions |
| Sanger Sequencing | >700 bp | Very long reads; low error rate; large insert sizes [37] | Small-scale studies where complete genomes from low-diversity environments are targeted |
Modern short-read platforms like Illumina provide extraordinary throughput and accuracy at continuously decreasing costs, making them suitable for large-scale metagenomic surveys [37] [1]. The technology uses sequencing-by-synthesis with reversible terminators, generating hundreds of millions of reads per run [37]. For applications requiring longer reads to improve assembly contiguity and resolve repetitive regions, emerging technologies like PacBio HiFi sequencing offer high-accuracy long reads that are particularly valuable for reconstructing metagenome-assembled genomes (MAGs) and performing strain-resolved analysis [33]. The DNBSEQ-G400 and DNBSEQ-T7 systems provide additional options for high-throughput metagenomic sequencing, with automated library preparation systems like the SP-960 streamlining the workflow for larger sample numbers [39].
Diagram 1: Library Preparation and Sequencing Workflow
Data Analysis and Functional Annotation
The analysis of shotgun metagenomic data involves multiple computational steps that transform raw sequencing reads into biological insights. A typical workflow begins with quality control and preprocessing, followed by taxonomic profiling, assembly, gene prediction, and functional annotation. The complexity of these analyses requires sophisticated bioinformatics tools and substantial computational resources.
Functional annotation is typically performed by comparing predicted protein-coding sequences against reference databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG) using BLASTP searches with specific thresholds (e.g., E value of ≤1e−5 and high-scoring segment pair score of >60) [40]. For each functional feature, abundance can be estimated by accumulating the relative abundance of all genes belonging to that feature, providing a quantitative view of the metabolic potential of the microbial community [40]. Advanced analysis pipelines like DRAGEN Metagenomics perform taxonomic classification of reads and provide both single-sample and aggregate reporting, facilitating comparative analyses across sample groups [1].
Diagram 2: Data Analysis Pipeline for Shotgun Metagenomics
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents and Tools for Shotgun Metagenomics
| Product Name | Function | Application Note |
|---|---|---|
| MGIEasy Stool Microbiome DNA Extraction Kit II [39] | Optimized DNA extraction from stool samples | Maximizes yield and quality from complex stool samples; available in 96-RXN and 384-RXN formats |
| DNBSEQ Fast FS Library Prep Kit V2.0 [39] | Preparation of sequencing libraries | Compatible with automated systems; suitable for high-throughput applications |
| FCL PE150 High-Throughput Sequencing Set [39] | Sequencing consumables | For paired-end 150 bp reads on DNBSEQ platforms |
| PFI Pro Software [39] | Bioinformatics analysis | Platform for microorganism identification and assembly evolution |
| ZTRON Lite Data Center [39] | Computational hardware | Dedicated system for handling large metagenomic datasets |
| Decoglurant | Decoglurant, CAS:911115-16-7, MF:C21H11F6N5, MW:447.3 g/mol | Chemical Reagent |
| Dersimelagon | Dersimelagon|Selective MC1R Agonist|RUO | Dersimelagon is an oral, selective MC1R agonist for research use only (RUO). It is investigated for photodermatoses and systemic sclerosis. Not for human use. |
This toolkit highlights essential reagents and platforms that support the entire workflow from sample preparation to data analysis. Automated systems such as the SP-100 Automated Sample Preparation System and SP-960 High-Throughput Automated Sample Preparation System can significantly improve reproducibility and throughput, especially for larger studies [39]. For specialized applications like strain-resolved analysis in inflammatory bowel disease or characterization of the "sexome" for forensic applications, PacBio HiFi metagenomic sequencing has demonstrated particular value by enabling precise functional gene profiling and capturing fine-scale microbial dynamics missed by short-read approaches [33].
Applications in Microbiome Functional Analysis Research
Shotgun metagenomics provides powerful applications for understanding microbiome function across diverse research areas. In human health, this approach has revealed how gut microbiomes contribute to conditions like inflammatory bowel disease (IBD) and childhood growth stunting, enabling researchers to move beyond taxonomic composition to understand functional capabilities [33]. The method also offers unique insights into microbial communities associated with specific body sites; for instance, skin microbiome studies have demonstrated clear functional differences between forehead (dominated by Cutibacterium spp.) and armpit (dominated by Staphylococcus spp.) communities [38].
In the realm of disease pathogenesis, shotgun metagenomics enables the investigation of functional roles played by microorganisms in conditions like colorectal cancer, where researchers can identify specific microbes involved in the adenoma-carcinoma sequence and potentially reveal novel targets for microbiome-based prevention and therapy [33]. The method's ability to reconstruct metagenome-assembled genomes (MAGs) from complex samples allows for strain-level analysis and the identification of microbial functions that may be associated with health and disease states, providing a more nuanced understanding of host-microbe interactions than what is possible with marker gene approaches alone.
Shotgun metagenomic sequencing has revolutionized microbiome research by enabling comprehensive analysis of the taxonomic and functional potential of microbial communities without the need for cultivation [41]. This approach involves sequencing random fragments of DNA from an environmental sample, followed by sophisticated bioinformatic processing to answer two fundamental questions: "Who is there?" and "What are they doing?" [42]. The bioinformatic workflow for shotgun metagenomics is complex and multi-stage, requiring careful selection of tools and parameters at each step to generate meaningful biological insights. This protocol details the essential computational steps for processing raw sequencing reads into annotated metagenome-assembled genomes (MAGs) and functional profiles, with particular emphasis on applications in human health and drug development research. The workflow is particularly valuable for exploring the functional capabilities of microbial communities in diverse environments, from the human gut to industrial settings [43] [44].
The standard bioinformatic workflow for shotgun metagenomics consists of four primary stages: quality control and preprocessing, assembly, binning, and gene annotation. Each stage employs specialized computational tools and generates specific output metrics for quality assessment. The following diagram illustrates the complete workflow and the relationships between each processing stage:
Quality Control and Preprocessing
Protocol Objectives
The initial quality control (QC) stage ensures that downstream analyses are performed on high-quality data by identifying sequencing artifacts, removing low-quality bases, and eliminating contaminating host DNA. This critical first step significantly impacts all subsequent analyses, as errors introduced during sequencing can compromise assembly and binning results [42].
Experimental Procedures
Quality Assessment with FastQC
Begin by assessing raw read quality using FastQC. Execute the following command for each sample:
This generates HTML reports containing key metrics including per-base sequence quality, sequence duplication levels, adapter content, and GC distribution. Examine these reports to identify potential issues requiring remediation in the trimming step [42].
Read Trimming and Filtering
Use Trimmomatic to remove adapters and low-quality bases:
This command performs the following operations: removes Illumina adapters with maximum mismatch count of 2; removes leading and trailing bases with quality below 3; scans reads with a 4-base sliding window, cutting when average quality per base drops below 15; and discards reads shorter than 50 bases [42].
Host DNA Removal
When working with samples containing substantial host DNA (e.g., human skin or gut samples), align reads to the host reference genome using BWA and retain only unmapped reads:
This process is particularly crucial for low-biomass samples like those from skin environments, where host contamination can substantially reduce microbial sequencing depth [38].
Quality Assessment Metrics
After processing, verify QC success by re-running FastQC on the cleaned reads and comparing reports. Key metrics indicating successful QC include: Q scores ≥30 across most bases, minimal adapter contamination, and appropriate GC content distribution for the sample type.
Metagenome Assembly
Protocol Objectives
Metagenome assembly reconstructs contiguous DNA sequences (contigs) from overlapping sequencing reads, providing longer fragments that facilitate more accurate gene prediction, taxonomic classification, and genome binning. The complexity of microbial communities and presence of strain-level variation make metagenome assembly computationally challenging [44].
Experimental Procedures
Assembly with metaSPAdes
Assemble quality-controlled reads using metaSPAdes, which is specifically designed for metagenomic data:
This command utilizes 16 threads (-t 16) and 200GB of memory (-m 200). Adjust these parameters based on available computational resources. metaSPAdes employs a multi-sized de Bruijn graph approach that effectively handles varying coverage levels across different microbial species in the community [44].
Assembly Quality Assessment
Evaluate assembly quality by examining contig statistics:
Key metrics to review include: N50 (contig length at which 50% of the assembly is contained in contigs of that length or longer), largest contig size, total assembly length, and number of contigs. Higher N50 values generally indicate better assembly, facilitating improved binning results [44].
Application Notes
For complex microbial communities or samples with highly uneven species abundance, consider co-assembly of multiple related samples to increase coverage for rare species. However, this requires significant computational resources and may introduce chimeric contigs from closely related strains [44].
Binning and Genome Reconstruction
Protocol Objectives
Binning clusters contigs into groups representing individual microbial genomes (Metagenome-Assembled Genomes or MAGs) based on sequence composition and abundance patterns across samples. This process enables species- or strain-level analysis of uncultivated microorganisms and is essential for associating specific functions with their microbial hosts [44].
Computational Methodologies
Composition-Based Binning
Composition-based methods utilize inherent genomic signatures such as k-mer frequencies (typically 4-mers), GC content, and codon usage. These approaches assume that sequences from the same genome share similar compositional patterns, while sequences from different genomes exhibit distinct signatures [45] [44].
Abundance-Based Binning
Abundance-based methods leverage coverage information across multiple samples, operating on the principle that contigs from the same genome exhibit similar abundance profiles across different samples. This approach is particularly effective for distinguishing closely related strains with similar sequence compositions but different abundance patterns [44].
Hybrid Binning Implementation
Execute binning using MetaBAT2, which combines compositional and abundance features:
For improved results, implement a hybrid approach using metaWRAP, which combines multiple binning tools:
Performance Evaluation of Binning Tools
Table 1: Comparison of Genome Binning Tools Based on Chicken Gut Metagenomic Datasets
| Tool | Type | Number of Bins | Completeness | Purity | Use Case |
|---|---|---|---|---|---|
| MetaBAT2 | Original | Medium | High | Medium | General purpose |
| GroopM2 | Original | Medium | High | High | High-purity requirements |
| MaxBin2 | Original | Medium | Medium | Medium | General purpose |
| Vamb | Original | High | Medium | High | Large datasets |
| AutoMeta | Original | Medium | High | Medium | Automated processing |
| metaWRAP | Refiner | Medium | High | High | Combining multiple binners |
| DAS Tool | Refiner | Medium | High | High | Combining multiple binners |
Evaluation data derived from testing on chicken gut metagenomic datasets demonstrates that MetaBAT2, GroopM2, and Autometa generally perform better than other original binners, while metaWRAP combining multiple binners produces the highest-quality genome bins [44].
Quality Assessment of MAGs
Assess the quality of reconstructed MAGs using CheckM, which evaluates completeness and contamination based on the presence of single-copy marker genes:
The Minimum Information about a Metagenome-Assembled Genome (MIMAG) standard recommends that high-quality MAGs should exhibit >90% completeness and <5% contamination, while medium-quality MAGs should have >50% completeness and <10% contamination [44].
Gene Prediction and Functional Annotation
Protocol Objectives
Functional annotation identifies genes within MAGs and contigs, then assigns putative functions based on homology to known proteins. This process enables metabolic reconstruction and functional profiling of microbial communities, moving beyond taxonomic composition to understand the biochemical capabilities of the microbiome [40] [46].
Experimental Procedures
Gene Prediction
Predict protein-coding genes on MAGs and unbinned contigs using Prodigal:
The -p meta parameter enables metagenomic mode, which is optimized for short, anonymous coding sequences without training on specific genomes.
Functional Annotation with KEGG
Annotate predicted proteins using the KEGG database via BLASTP search:
For each KEGG Orthology (KO) functional feature, estimate abundance by summing the relative abundance of all genes assigned to that feature [40]. Use an E-value cutoff of ≤1e−5 and a high-scoring segment pair (HSP) score of >60 to ensure annotation quality.
Annotation with MG-RAST
As an alternative to local annotation, upload sequences to MG-RAST for automated processing:
- Create an account at https://mg-rast.org
- Upload assembled contigs or MAGs in FASTA format
- Provide appropriate metadata following GSC standards
- Select quality control options including artificial duplicate read removal and quality-based trimming
- Submit for automated annotation against multiple databases including SEED, KEGG, and COG [46]
Advanced Functional Analysis
Metabolic Pathway Reconstruction
The transition from "bag-of-genes" to "bag-of-genomes" enables more accurate metabolic reconstruction by associating genes with their genomic context [43]. Use pathway tools such as MetaCyc or KEGG Mapper to visualize reconstructed metabolic pathways from KO assignments.
Genome-Scale Metabolic Modeling
For advanced functional analysis, construct genome-scale metabolic models from high-quality MAGs using tools like CarveMe or ModelSEED. These models enable computational simulation of metabolic fluxes and prediction of community interactions [43].
The Scientist's Toolkit
Table 2: Essential Research Reagent Solutions for Shotgun Metagenomics
| Tool/Resource | Function | Application Context |
|---|---|---|
| FastQC | Quality control assessment | Visualizes base quality scores, GC content, adapter contamination in raw reads |
| Trimmomatic | Read trimming | Removes adapters and low-quality bases using sliding window approach |
| metaSPAdes | Metagenome assembly | Constructs contigs from reads using de Bruijn graphs optimized for metagenomes |
| MetaBAT2 | Genome binning | Clusters contigs into MAGs using composition and abundance features |
| CheckM | MAG quality assessment | Evaluates completeness and contamination using single-copy marker genes |
| Prodigal | Gene prediction | Identifies protein-coding genes in prokaryotic sequences |
| KEGG Database | Functional annotation | Provides KO terms for metabolic pathway reconstruction |
| MG-RAST | Automated annotation pipeline | Web service for quality control, annotation, and comparative analysis |
| Diethyl-pythiDC | Diethyl-pythiDC, MF:C14H14N2O4S, MW:306.34 g/mol | Chemical Reagent |
| Dinordrin | Dinordrin, CAS:64675-08-7, MF:C27H36O4, MW:424.6 g/mol | Chemical Reagent |
Troubleshooting and Optimization
Low Assembly Quality
If assembly statistics show poor N50 values and fragmented contigs, consider: increasing sequencing depth, using longer read technologies (e.g., PacBio HiFi), or adjusting k-mer sizes in the assembler. For low-biomass samples like skin, optimize DNA extraction protocols to maximize microbial DNA yield [38].
Poor Binning Resolution
When binning produces highly contaminated or fragmented MAGs: implement hybrid binning approaches combining multiple tools, increase sample number to improve abundance profiling, or apply refinement tools like DAS Tool to consolidate results from different binners [44].
Limited Functional Annotation
For samples with novel microorganisms showing limited homology to database sequences: incorporate structural annotation of non-coding RNAs, use domain-based annotation tools (e.g., HMMER) with protein family models, or employ unsupervised gene clustering to identify novel gene families [47].
This protocol provides a comprehensive framework for bioinformatic processing of shotgun metagenomic data, from raw sequencing reads to functional annotations. The workflow enables researchers to reconstruct Metagenome-Assembled Genomes and characterize their functional potential, supporting advanced analyses in microbiome research. As the field evolves, integration of long-read sequencing, improved binning algorithms, and more sophisticated metabolic modeling will further enhance our ability to decipher the functional capabilities of microbial communities. Proper implementation of these bioinformatic procedures forms the foundation for robust microbiome research with applications in human health, biotechnology, and drug development.
Shotgun metagenomic sequencing has revolutionized microbiome research by enabling comprehensive analysis of the genetic potential of microbial communities. Unlike targeted amplicon sequencing, which identifies which microorganisms are present, shotgun sequencing reveals what functions these communities are capable of performing by sequencing all genomic DNA in a sample [2]. This approach provides unparalleled insights into functional potential, metabolic pathways, and microbial activities that influence host health and disease states.
The analysis of microbial metabolic pathways represents a critical frontier in understanding how microbiomes contribute to systems ranging from human health to environmental processes. By moving beyond taxonomic census to functional characterization, researchers can identify specific microbial genes, enzymes, and metabolic networks that drive ecosystem functions. This application note provides a structured framework for conducting these analyses, with detailed protocols and resources for profiling microbial metabolic pathways using shotgun metagenomic data.
Current Research and Applications
Recent investigations demonstrate the power of functional metagenomics across diverse fields. In 2025, several groundbreaking studies have received recognition for their innovative approaches to decoding microbial metabolism:
Table 1: Recent Research Applications of Functional Metagenomics
| Research Focus | Key Metabolic Findings | Technical Approach |
|---|---|---|
| Childhood Malnutrition [33] | Gut microbiome differences linked to growth stunting; intergenerational microbial patterns | HiFi shotgun metagenomics of mother-child dyads |
| Inflammatory Bowel Disease [33] | Precise functional gene profiling and strain-resolved analysis in IBD | PacBio-compatible protocols; HUMAnN 4 for functional profiling |
| Colorectal Cancer [33] | Microbial metabolic pathways associated with adenoma-carcinoma sequence | HiFi metagenomics for Metagenome-Assembled Genomes (MAGs) |
| Diet-Microbiome Interactions [48] | Enhanced pathways of tryptophan, galactose, fructose, and mannose metabolism after dietary shift | Multi-omics (shotgun metagenomics, LC-MS/MS metabolomics) |
| Antibiotic Resistance [49] | Elevated carbohydrate metabolism in fungal-dominated microbiomes; distinct ARG profiles | Illumina shotgun sequencing; CARD database for ARG identification |
| Wastewater Treatment [50] | Microbial co-metabolic degradation of organic micropollutants via specific enzymes | Integrated multi-omics (metagenomics, metaproteomics, metabolomics) |
| Myocardial Infarction [51] | Dysregulated glycerolipid metabolism and mTOR signaling pathways linked to blood microbiome | 16S rRNA sequencing integrated with LC-MS metabolomics |
These studies collectively highlight the methodological diversity and translational potential of functional metagenomic approaches. The integration of shotgun metagenomics with other omics technologies has been particularly valuable for connecting microbial taxa to specific metabolic functions and health outcomes.
Experimental Workflow for Functional Metagenomics
A robust experimental workflow is essential for generating high-quality functional metagenomic data. The following diagram illustrates the comprehensive pipeline from sample collection to functional interpretation:
Sample Collection and Preservation
Proper sample handling is critical for preserving microbial DNA and ensuring representative community analysis:
- Sterile Collection: Use sterile containers and instruments to prevent contamination from external microbes [2].
- Temperature Control: Freeze samples immediately after collection at -20°C or -80°C. Snap-freezing in liquid nitrogen is optimal for preserving DNA integrity [2].
- Preservation Buffers: When immediate freezing isn't possible, use preservation buffers (e.g., RNAlater) to maintain sample integrity for hours to days before freezing [2].
- Consistency: Maintain consistent collection methods across all samples to minimize technical variability [2].
For blood microbiome studies, additional precautions are essential: all reagents and materials must be meticulously sterilized, and certified medical personnel should adhere to strict protective protocols during collection to prevent contamination [51].
DNA Extraction and Library Preparation
The DNA extraction process significantly influences microbial community representation:
- Lysis Optimization: Combine chemical (enzymatic) and mechanical (bead-beating) methods to ensure complete cell lysis across diverse microbial taxa [2].
- Inhibitor Removal: Implement additional purification steps to remove contaminants like humic acids (soil samples) or host compounds (blood/tissue samples) [2] [51].
- Quality Assessment: Quantify DNA using fluorometric methods (e.g., Qubit dsDNA HS Assay) rather than spectrophotometry for accurate measurement of double-stranded DNA [51].
- Library Construction: Fragment DNA to 350-500bp, ligate index adapters for sample multiplexing, and clean up libraries to ensure proper size selection [40].
For functional studies, the absence of PCR amplification in shotgun metagenomics avoids primer bias and PCR artifacts, providing more accurate representation of gene abundances [2].
Sequencing Considerations
Sequencing platform and depth significantly impact functional analysis:
- Platform Selection: Illumina platforms provide high throughput for community-level profiling, while PacBio HiFi sequencing offers long reads for improved genome assembly and strain resolution [33].
- Sequencing Depth: Aim for 10-20 million reads per sample for standard functional profiling. Deeper sequencing (30-50 million reads) is required for metagenome assembly and rare gene detection [2].
- Controls: Include negative (extraction) controls and positive controls (mock communities) to monitor contamination and assess technical variability [2].
Computational Analysis of Metabolic Pathways
The computational workflow for functional analysis involves multiple steps to translate raw sequencing data into biological insights. The following diagram illustrates the key analytical steps and their relationships:
Quality Control and Preprocessing
Raw sequencing data requires rigorous preprocessing before functional analysis:
- Quality Filtering: Use Trimmomatic to remove low-quality bases and adapter sequences [51].
- Host DNA Removal: Align reads to host reference genomes (e.g., human, mouse) and remove matching sequences to enrich for microbial DNA [52].
- Error Correction: Apply algorithms to correct sequencing errors that might affect downstream annotation.
Tools like KneadData and BBDuk provide integrated solutions for quality control and host removal in metagenomic datasets.
Functional Annotation and Profiling
Functional annotation assigns biological meaning to metagenomic sequences:
- Gene Calling: Predict open reading frames (ORFs) in assembled contigs or directly on reads using tools like Prodigal or FragGeneScan.
- Database Alignment: Annotate genes by alignment to functional databases using BLASTP or more rapid alternatives like DIAMOND [24].
- Orthology Assignment: Assign KEGG Orthology (KO) terms to identify conserved functions across taxa [40].
- Specialized Annotation: Identify specific enzyme classes like CAZymes (carbohydrate-active enzymes) using dbCAN3 [24] or antibiotic resistance genes (ARGs) using CARD [49].
Table 2: Key Databases for Functional Annotation
| Database | Primary Use | Key Features | Annotation Tool |
|---|---|---|---|
| KEGG [40] [50] | Metabolic pathway annotation | Comprehensive pathway maps, KO hierarchy | KofamScan, BlastKOALA |
| CAZy [24] | Carbohydrate-active enzymes | Family classification of enzymes | dbCAN3 |
| CARD [49] | Antibiotic resistance genes | Comprehensive resistance ontology | RGI, DeepARG |
| GO | Gene ontology | Standardized biological processes | InterProScan |
| eggNOG [24] | Orthologous groups | Functional classification | eggNOG-mapper |
Metabolic Pathway Analysis
Advanced methods for pathway analysis provide systems-level understanding of microbial metabolism:
- Pathway Abundance Estimation: Tools like HUMAnN3 use a tiered approach to map reads to protein families (UniRef90) and reconstruct pathway abundances [52].
- Metagenomic Species Pan-genomes: Meteor2 leverages environment-specific gene catalogs to profile 11,653 metagenomic species pangenomes (MSPs) annotated with KEGG orthology, CAZymes, and antibiotic resistance genes [24].
- Metabolic Modeling: Genome-scale metabolic models (GEMs) enable in-silico simulation of metabolic networks, predicting microbial growth, nutrient utilization, and metabolite production [53].
- Strain-Level Analysis: Tools like StrainPhlAn detect single nucleotide variants in marker genes to track strain dissemination and functional variation [24].
For comprehensive analysis, Meteor2 has demonstrated strong performance in integrated taxonomic, functional, and strain-level profiling (TFSP), improving species detection sensitivity by 45% and functional abundance estimation accuracy by 35% compared to previous tools [24].
Integrated Multi-omics Approaches
Combining metagenomics with other data types provides a more complete picture of microbial metabolic activity:
Metagenomics-Metabolomics Integration
Integrated analysis of microbial genes and metabolites reveals active biological processes:
- Correlation Analysis: Statistical correlation of microbial abundance with metabolite levels to identify potential producer-consumer relationships [48].
- Pathway Mapping: Joint visualization of enriched metabolic pathways from both metagenomic and metabolomic data [48].
- Machine Learning: Application of random forests or other algorithms to identify microbiome-metabolite biomarkers of disease states [48] [51].
In diet-microbiome studies, this approach has revealed how transitions from high-protein to high-fiber diets enhance microbial diversity and promote specific metabolic pathways including tryptophan, galactose, fructose, and mannose metabolism [48].
Metaproteomics and Metatranscriptomics
- Functional Validation: Metaproteomics confirms which predicted genes are actually translated into proteins, validating functional predictions from metagenomics [50].
- Activity Assessment: Metatranscriptomics reveals which genes are actively transcribed under different conditions, providing insights into regulatory responses.
In wastewater treatment studies, integrating metagenomics with metaproteomics identified key microbial enzymes (oxidoreductases, hydrolases) involved in co-metabolic degradation of organic micropollutants [50].
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions for Functional Metagenomics
| Reagent/Kit | Function | Application Notes |
|---|---|---|
| TGuide S96 Magnetic Soil/Stool DNA Kit [51] | Microbial DNA extraction from complex samples | Optimized for difficult-to-lyse microbes; suitable for blood, soil, and stool |
| Agencourt AMPure XP Beads [51] | PCR amplicon purification | Size selection and cleanup of sequencing libraries |
| Qubit dsDNA HS Assay Kit [51] | Accurate DNA quantification | Fluorometric measurement of double-stranded DNA |
| NovaSeq 6000 Reagents [51] | High-throughput sequencing | Provides massive sequencing capacity for complex metagenomes |
| PacBio SMRTbell Libraries [33] | Long-read sequencing preparation | Enables HiFi sequencing for improved assembly and strain resolution |
| KOD FX Neo Polymerase [51] | High-fidelity PCR amplification | Minimal error rate for amplification of marker genes |
| Dipraglurant | Dipraglurant|mGlu5 NAM|For Research | Dipraglurant is a potent, selective mGlu5 negative allosteric modulator (NAM) for Parkinson's disease and dyskinesia research. For Research Use Only. Not for human use. |
| Diprovocim | Diprovocim |
Troubleshooting and Optimization
Common challenges in functional metagenomics and their solutions:
- Low Microbial DNA Yield: Add mechanical lysis steps (bead-beating) to DNA extraction protocol; increase starting sample volume; use carrier RNA during precipitation [2].
- Host DNA Contamination: Implement gradient centrifugation or filtration to enrich for microbial cells; use oxidative stress to differentially lyse human cells [2].
- Database Limitations: Combine multiple databases for annotation; use assembly-based approaches to discover novel genes; consider custom database construction for specialized environments [2].
- Computational Resource Constraints: Utilize "fast mode" in tools like Meteor2 that uses signature genes for rapid profiling with minimal resources (5 GB RAM, 10 min processing time) [24].
Future Directions
Emerging methodologies are enhancing our ability to decipher microbial metabolism:
- Long-Read Sequencing: PacBio HiFi sequencing provides complete genomes from metagenomes, enabling more accurate gene calling and pathway reconstruction [33].
- Single-Cell Metagenomics: Resolution of microbial metabolism at the single-cell level reveals functional heterogeneity within communities.
- Machine Learning Integration: Prediction of metabolic interactions and community dynamics from complex metagenomic datasets.
- Personalized Medicine Applications: Functional metagenomics for precision nutrition and microbiome-based therapeutics targeting specific metabolic pathways [53].
Functional analysis of microbial metabolism through shotgun metagenomics provides powerful insights into the activities of complex microbial communities. The integrated approaches and detailed protocols outlined in this application note empower researchers to move beyond taxonomic characterization to understand the functional capabilities of microbiomes and their impacts on health, disease, and ecosystem functioning. As databases expand and computational methods improve, functional metagenomics will continue to reveal the intricate metabolic networks that underlie microbial community dynamics.
Antimicrobial resistance (AMR) poses a significant global health threat, with an estimated 1.27 million deaths annually attributed to drug-resistant infections [54]. The characterization of resistance genes within microbial communities, known as the "resistome," has been revolutionized by shotgun metagenomic sequencing. This culture-independent approach enables comprehensive profiling of antibiotic resistance genes (ARGs) directly from environmental, clinical, and agricultural samples, providing insights into the diversity, abundance, and dynamics of resistance elements within complex microbial ecosystems [49]. The resistome represents the complete repertoire of ARGs within a microbiome, and its profiling is essential for understanding the emergence and dissemination of resistance across One Health domains connecting humans, animals, and the environment [54].
The application of shotgun metagenomics to AMR research allows researchers to move beyond culturable organisms and investigate the vast resistance potential of diverse microbial communities. This approach has revealed that fungal-dominated environments exhibit distinct resistome profiles compared to bacterial-rich communities, with lower bacterial diversity correlating with reduced abundance of certain ARGs [49]. Furthermore, metagenomic surveillance has illuminated the critical role of mobile genetic elements, particularly plasmids, as primary vectors for the horizontal transfer of ARGs between diverse bacterial taxa, enabling rapid dissemination of resistance across taxonomic boundaries and ecological habitats [54] [55].
Standardized Workflows for Resistome Analysis
Sample Processing and DNA Sequencing
The initial phase of resistome profiling involves sample collection, nucleic acid extraction, and library preparation for high-throughput sequencing. While specific protocols vary based on sample type (e.g., fecal, soil, water), the following general principles apply:
- Sample Preservation: Immediate freezing at -80°C or use of stabilization buffers to prevent microbial community shifts
- DNA Extraction: Implement bead-beating or enzymatic lysis for comprehensive cell disruption across diverse taxa
- Quality Control: Assess DNA integrity via fluorometry and gel electrophoresis; ensure concentration >1ng/μL
- Library Preparation: Fragment DNA to 200-500bp, attach platform-specific adapters, and amplify via PCR
- Sequencing: Utilize Illumina, PacBio, or Oxford Nanopore platforms with minimum 10 million paired-end reads (2×150bp) for adequate coverage [56] [57]
Bioinformatics Pipelines for Resistome Characterization
Several integrated bioinformatics platforms have been developed to streamline resistome analysis from raw sequencing data. These tools facilitate end-to-end processing, from quality control to ARG identification and visualization.
Table 1: Bioinformatics Tools for Resistome Analysis
| Tool | Primary Function | ARG Database | Key Features |
|---|---|---|---|
| AMRViz [56] | End-to-end analysis & visualization | CARD, ResFinder | Pan-genome analysis, phylogenetics, resistance/virulence profiling |
| Meteor2 [57] | Taxonomic/functional/strain profiling | ResFinder, ResFinderFG | Environment-specific gene catalogs, strain-level SNP tracking |
| AMRomics [56] | Pipeline integration | Multiple databases | Variant calling, pan-SNP analysis, reference-free workflow |
| R Packages [58] | Statistical analysis & visualization | Custom databases | Diversity analysis, differential abundance, correlation networks |
The following workflow diagram illustrates the standard bioinformatics pipeline for resistome analysis:
Analysis of Resistome Dynamics and Horizontal Gene Transfer
Taxonomic and Functional Profiling of Resistomes
Shotgun metagenomics enables simultaneous characterization of taxonomic composition and functional potential, including ARG profiles. A comparative study of fungal-dominated (HFJ) versus bacterial-rich (QFJ) fermentation environments revealed stark contrasts in resistome composition [49]. The HFJ samples, dominated by Saccharomyces cerevisiae, exhibited elevated carbohydrate metabolism and reduced ARG abundance, while QFJ samples with higher bacterial diversity (particularly Firmicutes and Proteobacteria) showed enrichment in lipid and amino acid metabolism pathways and significantly greater ARG abundance, particularly genes conferring resistance to beta-lactams, aminoglycosides, and tetracyclines [49].
Table 2: Comparative Resistome Profiles Across Microbial Ecosystems
| Sample Type | Dominant Taxa | Primary Metabolic Pathways | ARG Abundance | Predominant ARG Classes |
|---|---|---|---|---|
| Fungal-rich (HFJ) [49] | Eukaryota (S. cerevisiae) | Carbohydrate metabolism | Low | Limited diversity |
| Bacterial-rich (QFJ) [49] | Firmicutes, Proteobacteria | Lipid & amino acid metabolism | High | Beta-lactams, aminoglycosides, tetracyclines |
| K. pneumoniae clinical isolates [55] | Proteobacteria | Diverse virulence factors | High | Carbapenems, multidrug efflux |
Plasmid-Mediated Resistance Dissemination
Plasmids serve as cornerstone elements in the global spread of AMR, facilitating the horizontal transfer of ARGs across taxonomic boundaries and between habitats [54]. Recent research has highlighted the particular significance of mobilizable plasmids, which were previously underestimated in their contribution to resistance dissemination. Unlike self-transmissible conjugative plasmids that encode complete transfer machinery, mobilizable plasmids lack partial conjugative elements and transfer with helper conjugative plasmids [55].
Studies in Klebsiella pneumoniae have demonstrated that mobilizable multidrug-resistant (MDR) and virulence plasmids carrying the origin of transfer (oriT) but lacking relaxase genes can cotransfer with helper conjugative carbapenem-resistant plasmids across various Klebsiella and Escherichia coli strains [55]. Notably, these mobilizable plasmids carry approximately 20% of acquired antimicrobial resistance genes and more than 75% of virulence genes in K. pneumoniae, presenting a substantial threat in clinical settings [55]. Additionally, mobilizable plasmids exhibit enhanced stability in recipient strains and can evade CRISPR-Cas defense systems that typically target conjugative plasmid elements, providing them with a selective advantage in bacterial populations [55].
The following diagram illustrates the mechanisms of plasmid-mediated horizontal gene transfer:
Experimental Protocols for Resistome Tracking
Metagenomic Sequencing and Assembly Protocol
Objective: Comprehensive profiling of resistome composition and dynamics from complex microbial samples.
Materials:
- Extracted metagenomic DNA (>1ng/μL)
- Illumina-compatible library preparation kit
- Qubit fluorometer or equivalent
- Bioanalyzer/TapeStation for quality control
- High-performance computing cluster with minimum 16GB RAM
Procedure:
- Library Preparation:
- Fragment 100ng DNA to 300-500bp using acoustic shearing
- Repair ends, add A-overhangs, and ligate with indexed adapters
- Clean up with AMPure XP beads and amplify with 8 PCR cycles
- Validate library size distribution using Bioanalyzer
Sequencing:
- Pool libraries at equimolar concentrations
- Sequence on Illumina platform (NovaSeq 6000 recommended)
- Generate minimum 10 million 150bp paired-end reads per sample
Bioinformatics Processing:
- Quality control: Remove adapters and low-quality bases using Trimmomatic
- Metagenomic assembly: Utilize SPAdes (Illumina) or Flye (long-read) with metagenomic mode
- Gene prediction: Annotate contigs with Prokka for ORF identification
- ARG screening: Query predicted genes against CARD database using RGI
- Abundance quantification: Map reads to ARG database using Bowtie2 [56] [57]
Plasmid Transfer and Stability Assay
Objective: Evaluate conjugative transfer of mobilizable plasmids carrying ARGs and their stability in transconjugants.
Materials:
- Donor strain: K. pneumoniae HS11286 carrying conjugative (pHSKP2) and mobilizable (pHSKP3) plasmids
- Recipient strains: E. coli C600 (rifamycin-resistant) or K. variicola KvBSI002A (apramycin-resistant)
- LB broth and agar plates with selective antibiotics
- Nitrocellulose filters (0.45μm pore size)
Procedure:
- Bacterial Culture:
- Inoculate separate overnight cultures of donor and recipient strains in LB broth with appropriate antibiotics
- Subculture 1:100 in fresh LB and incubate at 37°C with shaking until OD600 reaches 0.6
Conjugation Assay:
- Mix 1mL each of donor and recipient cells, wash with PBS, and resuspend in 20μL 10mM MgSO₄
- Transfer to nitrocellulose filter on pre-warmed LB agar plate
- Incubate overnight at 37°C
Transconjugant Selection:
- Resuspend cells in PBS and plate serial dilutions on selective media:
- E. coli transconjugants: LB + rifamycin (50μg/mL) + meropenem (0.2μg/mL) for CR plasmid or kanamycin (50μg/mL) for MDR plasmid
- K. variicola transconjugants: LB + apramycin (50μg/mL) + appropriate antibiotics
- Include appropriate controls (donor and recipient alone)
- Resuspend cells in PBS and plate serial dilutions on selective media:
Confirmation and Stability Testing:
- Verify transconjugants via replica plating and PCR with plasmid-specific primers
- Analyze plasmid morphology via XbaI and S1 nuclease PFGE
- Assess plasmid stability via serial passage culture without selective pressure
- Monitor plasmid retention over 10 generations by replica plating and PCR screening [55]
Research Reagent Solutions for Resistome Studies
Table 3: Essential Research Reagents and Computational Tools for Resistome Analysis
| Category | Specific Tool/Reagent | Application | Key Features |
|---|---|---|---|
| Sequencing Platforms | Illumina NovaSeq 6000 | High-throughput sequencing | 150bp paired-end reads, high coverage |
| Oxford Nanopore PromethION | Long-read sequencing | Structural variant detection, plasmid assembly | |
| Bioinformatics Tools | AMRViz [56] | Integrated analysis & visualization | Pan-genome analysis, phylogenetic tracking |
| Meteor2 [57] | Taxonomic/functional profiling | Environment-specific catalogs, strain-level resolution | |
| R Microbiome Packages [58] | Statistical analysis | Diversity metrics, differential abundance testing | |
| Reference Databases | CARD [49] | ARG annotation | Comprehensive resistance ontology |
| ResFinder [57] | ARG identification | Clinically relevant resistance determinants | |
| GTDB [57] | Taxonomic classification | Standardized microbial taxonomy | |
| Culture Resources | K. pneumoniae strains [55] | Conjugation assays | Well-characterized plasmid hosts |
| Selective antibiotics | Transconjugant selection | Meropenem, kanamycin, rifamycin, apramycin |
Advanced Applications and Integrative Approaches
Machine Learning in Resistome Analysis
Emerging data-driven approaches are enhancing our ability to predict and understand AMR patterns. Unsupervised learning techniques, including K-means clustering and principal component analysis (PCA), have been applied to identify inherent patterns in AMR gene data without predefined labels [59]. Analysis of the PanRes dataset, which synthesizes comprehensive data on AMR genes from multiple genomic databases, has revealed distinct clusters of AMR genes based on gene length and resistance class associations [59]. These patterns provide novel insights into potential resistance mechanisms, particularly the role of gene length in different resistance pathways, and can inform the development of predictive models for clinical resistance phenotypes.
Diagnostic and Therapeutic Implications
The World Health Organization's 2025 reports highlight critical gaps in the antibacterial development pipeline and diagnostic tools for AMR [60] [61]. The clinical antibacterial pipeline has decreased from 97 agents in 2023 to 90 in 2025, with only 15 classified as innovative and merely five effective against WHO "critical" priority pathogens [60] [61]. Simultaneously, diagnostic gaps persist, particularly in low-resource settings, including the absence of multiplex platforms for bloodstream infection identification and limited point-of-care tools for primary care facilities [60]. Resistome profiling through shotgun metagenomics offers promising approaches to address these challenges by enabling rapid identification of resistance patterns and informing targeted therapeutic strategies.
Integrated resistome analysis platforms like AMRViz and Meteor2 facilitate the translation of genomic data into clinically actionable insights by providing comprehensive visualization of resistance profiles, phylogenetic relationships, and virulence factors [56] [57]. These tools enable researchers and clinicians to track the dissemination of high-risk clones and resistance elements across healthcare settings and geographical regions, supporting evidence-based interventions and antimicrobial stewardship programs. As resistome profiling technologies continue to advance, they hold significant potential for guiding the development of novel antibacterial agents and precision medicine approaches to combat the escalating threat of antimicrobial resistance.
Microbial communities represent one of the most promising yet underexplored resources for novel therapeutic compounds. The historical approach to antibiotic discovery, which flourished between the 1940s and 1960s, has unfortunately reached a standstill, leaving our antibiotic pipeline critically limited despite the rising global threat of antimicrobial resistance (AMR) [62]. This challenge has catalyzed a paradigm shift from traditional cultivation-based methods toward innovative approaches that leverage next-generation sequencing and bioinformatics. Shotgun metagenomic sequencing has emerged as a transformative technology, enabling researchers to directly access the genetic blueprint of entire microbial communities without the need for cultivation [63] [37]. This approach provides unprecedented insight into the functional potential of microbiomes, opening new frontiers for discovering cryptic biosynthetic gene clusters (BGCs) that encode novel antimicrobial compounds, anticancer agents, and other therapeutic molecules [64].
The rationale for exploring microbial communities extends beyond mere convenience. Many human infections, including chronic cystic fibrosis (CF) lung disease and chronic non-healing wounds, are polymicrobial in nature [62]. In these complex communities, microbial interactions through metabolic cross-feeding, quorum sensing, and exchange of genetic material can dramatically alter pathogen susceptibility to antibacterial treatments [62]. Consequently, the traditional "one microbe, one disease" postulate and associated antimicrobial susceptibility testing (AST) methods often fail to predict treatment outcomes in clinical settings [62]. Shotgun metagenomics offers a pathway to overcome these limitations by providing a comprehensive view of microbial community structure, function, and interactions, thereby enabling more effective therapeutic strategies.
Shotgun Metagenomics in Drug Discovery: Workflow and Value Proposition
Comparative Analysis of Metagenomic Approaches
Table 1: Comparison of microbial community analysis approaches for drug discovery.
| Feature | 16S/18S/ITS Amplicon Sequencing | Shotgun Metagenomic Sequencing |
|---|---|---|
| Scope of Detection | Primarily bacteria and archaea (16S) or fungi (ITS) | All microbial domains: bacteria, archaea, viruses, fungi, protozoa |
| Taxonomic Resolution | Genus to species level | Species to strain level [65] |
| Functional Insights | Indirect inference via phylogenetic markers | Direct assessment of gene content and metabolic pathways [7] [63] |
| BGC Discovery Potential | Limited | High; enables identification of novel biosynthetic gene clusters [64] |
| PCR Amplification Bias | Present | Absent [2] |
| Relative Cost | Lower | Higher, though decreasing [2] |
Key Workflow in Drug Discovery
The application of shotgun metagenomics to drug discovery follows a structured pipeline from sample collection to compound identification. The critical stages include sample preparation, DNA sequencing, bioinformatic analysis, and experimental validation [63] [2]. Sample integrity is paramount, requiring strict adherence to sterility, immediate freezing at -20°C or -80°C, and minimization of freeze-thaw cycles to preserve an accurate representation of the microbial community [2]. Subsequent DNA extraction must be optimized for the sample type (e.g., soil, feces, tissue) to maximize yield and minimize contamination from inhibitors like humic acids [37].
Following library preparation, high-throughput sequencing generates billions of short DNA reads. Long-read sequencing technologies, such as PacBio SMRT systems, are particularly valuable for resolving complex genomic regions and assembling complete BGCs, which are often repetitive and difficult to reconstruct with short reads alone [63]. The resulting data then undergoes a rigorous bioinformatic analysis to answer two fundamental questions: "Who is there?" (taxonomic profiling) and "What are they doing?" (functional potential) [7]. This involves assembly, binning of contigs into metagenome-assembled genomes (MAGs), and annotation against functional databases [63] [37]. The identification of novel BGCs through genome mining strategies then guides the targeted isolation and structural elucidation of the encoded natural products [64] [66].
Figure 1: The end-to-end workflow for discovering novel therapeutic compounds from microbial communities using shotgun metagenomics.
Therapeutic Applications and Frontiers
Shotgun metagenomics is unlocking new therapeutic possibilities across multiple disease areas by providing strain-level resolution and functional insights previously unattainable.
Revitalizing Antibiotic Discovery
The decline in novel antibiotic discovery poses a grave threat to public health. Shotgun metagenomics addresses this by enabling the systematic mining of biosynthetic gene clusters from diverse microbiomes, including those from extreme environments and uncultured organisms [64]. This approach has revealed that sequenced microorganisms, particularly actinomycetes with large genomes, contain dozens of secondary metabolite BGCs that were not detected through traditional culture-based methods [64]. For instance, the discovery of Streptomyces coelicolor's true biosynthetic potential via genome sequencing demonstrated that even well-studied model organisms harbor untapped chemical diversity [64]. Furthermore, by studying polymicrobial communities, researchers can identify compounds that are only produced or become potent in the context of microbial interactions, such as chloroxylenol which shows significantly enhanced activity against S. aureus in the presence of P. aeruginosa metabolites [62].
Targeting the Microbiome in Human Disease
Strain-level microbiome sequencing is opening up several key therapeutic frontiers:
- Targeted Live Biotherapeutics: The 2023 FDA approval of SER-109, the first oral microbiome-based therapy for recurrent C. difficile infection, marks a paradigm shift toward "live" therapeutics [65]. The rational design and prescription of such therapies depend on precise strain-level characterization to ensure safety and efficacy [65].
- Uncovering Cancer-Linked Microbes: Specific microbial strains have been directly implicated in oncogenesis. Strain-level sequencing helps identify microbial signatures associated with cancers, such as colorectal and pancreatic cancer, suggesting that therapeutic breakthroughs may come from eliminating the triggering bacteria rather than targeting human mutations [65].
- Combatting Antimicrobial Resistance (AMR): Metagenomic sequencing allows for the comprehensive tracking of antibiotic resistance genes (e.g., using the CARD database) within microbiomes [63]. This insight is critical for understanding the emergence and spread of resistance and for developing targeted antibiotic stewardship strategies [65].
- Mapping the Gut-Brain Axis: Early research is beginning to link specific bacterial strains to mental health conditions like anxiety and depression [65]. Although an emerging field, this work hints at future opportunities for microbiome-targeted therapies for neuropsychiatric conditions.
Essential Protocols for Metagenome-Driven Drug Discovery
Protocol 1: Shotgun Metagenomic Sequencing for Biosynthetic Gene Cluster Discovery
Objective: To comprehensively profile the taxonomic and functional composition of a microbial community and identify putative biosynthetic gene clusters (BGCs) encoding novel therapeutic compounds.
Materials and Reagents: Table 2: Key research reagents and materials for shotgun metagenomic sequencing.
| Reagent/Material | Function | Example Kits/Platforms |
|---|---|---|
| Sample Preservation Buffer | Stabilizes microbial community DNA/RNA at ambient temperature | RNAlater, Zymo DNA/RNA Shield |
| DNA Extraction Kit | Lyses microbial cells and purifies high-molecular-weight DNA | DNeasy PowerSoil Pro Kit (QIAGEN) |
| Library Preparation Kit | Fragments DNA and adds sequencing adapters/barcodes | Illumina Nextera XT, PacBio SMRTbell |
| Sequencing Platform | Generates high-throughput sequence reads | Illumina NovaSeq, PacBio Sequel II, Ion Torrent |
| Bioinformatics Tools | Processes raw data for assembly, binning, and annotation | MG-RAST, HUMAnN, antiSMASH, MetaPhlAn [63] |
Methodology:
- Sample Collection and Preservation: Aseptically collect samples (e.g., soil, feces, water) using sterile instruments. Immediately snap-freeze in liquid nitrogen or preserve in an appropriate buffer. Store at -80°C until processing [37] [2].
- Metagenomic DNA Extraction: Extract DNA using a kit designed for the specific sample type. For soil or stool, this often involves mechanical lysis (bead beating) to ensure rupture of tough microbial cell walls. Quantify DNA yield using fluorometry and assess quality via agarose gel electrophoresis or Bioanalyzer [63] [2].
- Library Preparation and Sequencing: Fragment purified DNA to an appropriate size (e.g., 350-800 bp). Ligate sequencing adapters and sample-specific barcodes (multiplexing). For complex communities, aim for a minimum of 10-20 million reads per sample. Select the sequencing platform based on the need for read length (PacBio) versus high throughput (Illumina) [63] [37].
- Bioinformatic Analysis:
- Quality Control: Remove low-quality reads, adapter sequences, and host-derived reads (if applicable) using tools like Trimmomatic or KneadData.
- Assembly and Binning: Perform de novo assembly of quality-filtered reads using metaSPAdes or MEGAHIT. Group assembled contigs into Metagenome-Assembled Genomes (MAGs) based on sequence composition and abundance (binning) with tools like MaxBin or MetaBAT2 [63].
- Annotation and BGC Prediction: Annotate genes and pathways using databases such as KEGG, UniProt, and eggNOG [63]. Specifically scan for BGCs using the antiSMASH tool to identify regions encoding nonribosomal peptide synthetases (NRPS), polyketide synthases (PKS), and other secondary metabolite machinery [64].
Protocol 2: Functional Validation of Polymicrobial Community Interactions
Objective: To experimentally validate the hypothesis that microbial interactions within a polymicrobial community can modulate antibiotic tolerance and identify the underlying metabolites.
Background: Current Antimicrobial Susceptibility Testing (AST) fails to predict treatment outcomes for polymicrobial infections because it tests pathogens in isolation [62]. This protocol uses an in vitro model community to investigate these interactions.
Methodology:
- Design a Model Polymicrobial Community: Select microbial strains relevant to the disease context (e.g., for cystic fibrosis airways: Pseudomonas aeruginosa, Staphylococcus aureus, and Haemophilus influenzae). Start with a simplified 2-3 member community to ensure tractability [62].
- Cultivation in Disease-Relevant Media: Grow the community in a synthetic medium that mimics the in vivo environment (e.g., Synthetic Cystic Fibrosis Medium - SCFM2) rather than standard nutrient-rich broth. This promotes in vivo-like bacterial physiology [62] [37].
- Antibiotic Exposure and Metabolite Profiling:
- Treat the polymicrobial community and each monoculture control with a front-line antibiotic (e.g., vancomycin for S. aureus).
- Compare the minimum inhibitory concentration (MIC) and killing kinetics between the community and monoculture contexts.
- Collect cell-free supernatants from co-cultures and perform untargeted metabolomics (e.g., LC-MS) to identify metabolites exchanged between species that may confer antibiotic tolerance [62].
- Mechanistic Follow-up: Supplement monocultures with the identified metabolites and re-test antibiotic susceptibility to confirm their protective role.
Figure 2: Experimental workflow for validating polymicrobial interactions that influence antibiotic susceptibility. AST = Antimicrobial Susceptibility Testing.
The Scientist's Toolkit: Key Reagents and Databases
Successful implementation of the described protocols requires a suite of specialized reagents, tools, and databases.
Table 3: Essential bioinformatics databases and tools for metagenomic analysis and genome mining.
| Tool/Database | Primary Function | Application in Drug Discovery |
|---|---|---|
| antiSMASH | Identification & analysis of biosynthetic gene clusters (BGCs) | Core tool for predicting novel secondary metabolite pathways from MAGs [64] |
| KEGG | Annotation of genes and metabolic pathways | Understanding functional potential of community & identifying therapeutic targets [63] |
| CARD | Comprehensive Antibiotic Resistance Database | Profiling antibiotic resistance genes within the microbiome [63] |
| HUMAnN | Profiling abundance of microbial metabolic pathways | Quantifying community-wide functional differences between health and disease states [33] [63] |
| MG-RAST | Open-source metagenome analysis server | Accessible pipeline for annotation and comparative analysis [63] |
| MbtH Homolog Analysis | Beacon for counting NRPS pathways | Triage and prioritize "gifted" actinomycetes for further mining [64] |
| Diroximel Fumarate | Diroximel Fumarate|High-Purity Reference Standard | Diroximel fumarate is an immunomodulating agent for relapsing MS research. This product is for research use only (RUO) and not for human consumption. |
Shotgun metagenomic sequencing has fundamentally altered the landscape of microbial natural product discovery. By moving beyond the constraints of microbial cultivation and the limited resolution of amplicon sequencing, this powerful approach provides direct access to the immense genetic and functional diversity of microbial communities. The integration of strain-level profiling, functional gene analysis, and genome mining within a single framework enables researchers to not only catalog microbial inhabitants but also to uncover the precise genetic blueprints for novel therapeutic compounds. As sequencing technologies continue to advance and computational tools become more sophisticated, the drug discovery pipeline will be increasingly fueled by insights derived from microbiomes. This promises to revitalize the search for new antibiotics and open new therapeutic avenues in oncology, neuropsychiatry, and beyond, ultimately making precision microbiomics a foundational element of modern drug discovery and development.
The human gut microbiome, a complex ecosystem comprising trillions of microorganisms, has emerged as a critical factor influencing drug metabolism and treatment efficacy. This "second genome," harboring approximately 100 times more genes than the human host, possesses immense metabolic capacity that directly and indirectly modulates pharmaceutical compound activity [67]. Understanding these microbiome-drug interactions is fundamental to advancing precision medicine, as they contribute significantly to individual variability in drug response (IVDR) that cannot be fully explained by human genetic factors alone [67].
The gut microbiota influences drug pharmacokinetics and pharmacodynamics through multiple mechanisms, including direct enzymatic transformation of drugs, alteration of host metabolic pathways, modulation of immune responses, and competition for metabolic pathways with drug molecules [67]. These interactions can transform inactive prodrugs into their active forms, convert active drugs into toxic metabolites, or alter drug bioavailability, ultimately impacting both therapeutic efficacy and adverse drug reaction profiles [68]. Consequently, the burgeoning field of pharmacomicrobiomics aims to systematically characterize correlations between microbiota variation and drug response variability, offering promising avenues for optimizing therapeutic outcomes across diverse patient populations [67].
Analytical Approaches for Studying Microbiome-Drug Interactions
Shotgun Metagenomics for Comprehensive Profiling
Shotgun metagenomic sequencing has become the cornerstone technique for investigating microbiome-drug interactions at high resolution. Unlike targeted amplicon sequencing approaches that focus on specific taxonomic marker genes (e.g., 16S rRNA), shotgun metagenomics sequences all genetic material in a sample, enabling simultaneous assessment of taxonomic composition, functional potential, and strain-level variation within microbial communities [69]. This comprehensive approach is particularly valuable for pharmacomicrobiomics research as it provides direct access to the complete genetic repertoire of microbial communities, including genes encoding drug-metabolizing enzymes, antibiotic resistance determinants, and metabolic pathways that indirectly influence drug disposition [24] [69].
The analytical workflow for shotgun metagenomics involves multiple critical steps, each requiring careful optimization to ensure reproducible and biologically meaningful results. Key considerations include:
- Sample collection and DNA extraction: Standardized protocols are essential to minimize technical variability and potential biases introduced during these preliminary steps [69].
- Sequencing depth and strategy: Adequate sequencing depth (typically 10-14 Gb for complex gut samples) ensures sufficient coverage of low-abundance community members who may contribute disproportionately to drug metabolism [27].
- Bioinformatic processing: This involves quality control, host DNA depletion, assembly, gene prediction, and functional annotation using specialized databases and algorithms [24] [27].
- Multi-omic integration: Combining metagenomic data with metabolomic, transcriptomic, and proteomic profiles provides a more holistic understanding of functional activities and host-microbe interactions [70].
Table 1: Comparison of Microbiome Analysis Approaches
| Feature | Shotgun Metagenomics | Amplicon Sequencing |
|---|---|---|
| Scope | All genetic material in sample | Specific marker genes only |
| Taxonomic Resolution | Species and strain level | Genus level, limited species |
| Functional Insights | Direct assessment via gene content | Inferred from taxonomy |
| Cost per Sample | ~$1000+ | ~$50-100 |
| Computational Demand | High | Moderate |
| Drug Metabolism Applications | Identification of drug-metabolizing genes and pathways | Limited to correlating taxonomy with drug response |
Integrated Multi-Omics Frameworks
Advanced studies now employ integrated multi-omics frameworks that combine metagenomics with metabolomic profiling to establish mechanistic links between microbial genetic capacity and actual metabolic activities. For example, Ning et al. conducted a large-scale integration encompassing over 1,300 metagenomes and 400 metabolomes from inflammatory bowel disease patients and healthy controls, identifying consistent alterations in underreported microbial species alongside significant metabolite shifts that directly linked microbial community disruptions to disease status [70]. Similarly, Qin et al. employed high-resolution serum metabolomics to profile gut microbial composition and function in type 2 diabetes, identifying 111 gut microbiota-derived metabolites significantly associated with disease progression [70]. These integrated approaches demonstrate superior predictive power for disease diagnosis and stratification, with diagnostic models achieving area under the receiver operating characteristic curve values of 0.92-0.98 for distinguishing inflammatory bowel disease from controls [70].
Key Mechanisms of Microbiome-Drug Interactions
Direct Microbial Biotransformation of Drugs
Gut microorganisms express a diverse arsenal of enzymes that directly modify pharmaceutical compounds through various biotransformation reactions. These microbial transformations can profoundly alter drug pharmacokinetics, therapeutic efficacy, and toxicity profiles. The most significant reactions include:
Reductive Metabolism: Gut microbiota provide a major source of reductive metabolic capability, performing azo reduction, nitroreduction, and sulfoxide reduction [68]. The classic example is the reduction of the antibacterial prodrugs prontosil and neoprontosil to release active sulphanilamide [68]. Similarly, the therapeutic activity of 5-aminosalicylic acid prodrugs used in inflammatory bowel disease (e.g., sulfasalazine, olsalazine, balsalazide) depends entirely on microbial azo reduction in the gut [68]. Nitroreduction by gut bacteria also activates benzodiazepines containing nitro-groups (nitrazepam, clonazepam) to their amine metabolites, with studies demonstrating that antibiotic pretreatment in rats reduced nitroreduced metabolites from 30% to just 2% of excreted nitrazepam metabolites [68].
Hydrolytic Reactions: Gut microbiota extensively hydrolyze various drug conjugates, particularly glucuronides, through β-glucuronidase activity. This deconjugation can reactivate drugs previously detoxified by host conjugation pathways, potentially leading to localized gastrointestinal toxicity or enterohepatic recirculation [68] [67]. Additionally, hydrazone linkages in drugs like levosimendan and eltrombopag are susceptible to reductive cleavage by gut microbiota [68].
Additional Transformations: Beyond these major pathways, gut microbiota perform diverse other reactions including decarboxylation, dehydroxylation, demethylation, dehalogenation, dealkylation, and deamination [68]. For instance, the benzisoxazole ring of the anticonvulsant zonisamide undergoes reductive metabolism by several gut bacteria, with Clostridium sporogenes showing the highest activity [68].
Table 2: Clinically Relevant Direct Drug Metabolism by Gut Microbiota
| Drug Category | Example Drugs | Microbial Reaction | Metabolic Consequence |
|---|---|---|---|
| Prodrugs | Prontosil, Sulfasalazine | Azo reduction | Drug activation |
| Benzodiazepines | Nitrazepam, Clonazepam | Nitroreduction | Altered metabolism and toxicity |
| Anti-inflammatory | Sulindac | Sulfoxide reduction | Altered drug activity |
| Proton Pump Inhibitors | Omeprazole | Sulfoxide reduction | Metabolic switching |
| Cardiac Agents | Levosimendan | Hydrazone reduction | Cleavage and metabolite formation |
| Anticonvulsants | Zonisamide | Benzisoxazole reduction | Altered clearance and efficacy |
Indirect Modulation of Drug Disposition
Beyond direct biotransformation, gut microbiota indirectly influence drug disposition through multiple mechanisms:
Modulation of Host Drug Metabolism: Gut microbial metabolites, particularly short-chain fatty acids, secondary bile acids, and polyamines, can regulate the expression and activity of host hepatic drug-metabolizing enzymes (e.g., cytochrome P450 isoforms) and drug transporters [68] [67]. This microbial influence creates a complex interplay where microbiota-derived signals systematically alter the host's capacity to metabolize and eliminate pharmaceuticals.
Immune System Modulation: As a critical interface with the host immune system, gut microbiota shape systemic inflammatory tone and immune responses that subsequently influence drug targets and therapeutic outcomes, particularly for immunomodulatory agents and cancer therapeutics [67].
Competition for Metabolic Pathways: Microbiota and pharmaceutical compounds often compete for shared metabolic pathways, particularly conjugation reactions like glucuronidation and sulfation, potentially creating metabolic bottlenecks that alter drug clearance rates and metabolite patterns [67].
Experimental Protocols for Investigating Microbiome-Drug Interactions
Sample Collection and Metagenomic Sequencing
Materials:
- Sterile fecal collection containers with DNA stabilization buffer or rectal swab kits
- MP-soil FastDNA Spin Kit for Soil (MP Biomedicals, #6560-200) or equivalent
- Illumina, PacBio, or Oxford Nanopore sequencing platforms
- Host DNA depletion reagents (e.g., NEBNext Microbiome DNA Enrichment Kit)
Protocol:
- Sample Collection: Collect fresh fecal samples in sterile containers with DNA/RNA stabilization buffer, or obtain rectal swabs following standardized protocols [27]. For swabs, clean the perianal area with soap, water, and 70% alcohol, allow disinfectant evaporation, insert a sterile saline-moistened swab 4-5 cm into the anus, rotate gently, and immediately place in sterile tubes [27]. Flash-freeze samples in liquid nitrogen and store at -80°C.
- DNA Extraction: Perform DNA extraction using the MP-soil FastDNA Spin Kit according to manufacturer instructions [27]. Assess DNA purity, concentration, and quality using NanoDrop, fluorometry, and agarose gel electrophoresis.
- Library Preparation and Sequencing: Prepare metagenomic libraries using Illumina-compatible kits (e.g., Nextera XT) following manufacturer protocols. For functional analyses requiring longer reads, consider PacBio HiFi or Oxford Nanopore platforms [33]. Sequence on Illumina HiSeq 4000 or equivalent platforms to achieve minimum 10-14 Gb reads per sample [27].
- Quality Control: Process raw sequencing reads through fastp (v0.23.0) or similar tools to remove adapters, trim low-quality bases (quality score <20), and filter short reads (<50 bp) [27]. Remove human genomic reads by mapping to the human reference genome using BWA (v0.7.17) [27].
Metagenomic Data Analysis with Meteor2
Materials:
- High-performance computing cluster (≥5 GB RAM per sample)
- Meteor2 software (open source, available from original publication)
- Meteor2 database (includes 10 ecosystem-specific gene catalogues)
Protocol:
- Software Installation: Install Meteor2 following developer documentation. Download and configure the appropriate environment-specific microbial gene catalogue (human gut, oral, skin, mouse gut, etc.) [24].
- Taxonomic, Functional, and Strain-level Profiling (TFSP): Process quality-controlled reads through Meteor2 using default parameters (95% identity threshold for alignments) [24]. For rapid analysis, use "fast mode" with more stringent 98% identity threshold [24].
- Gene Counting and Normalization: Employ Meteor2's shared counting mode (default) where reads with multiple alignments contribute to gene counts based on proportion weights relative to each gene's unique read count [24]. Normalize gene counts using depth coverage (read count divided by gene length × 100) or FPKM [24].
- Taxonomic Profiling: Generate metagenomic species pangenome profiles by averaging abundance of signature genes (default: 100 most central genes per MSP) after normalization [24]. Apply detection threshold of 10% signature genes for full mode or 20% for fast mode [24].
- Functional Profiling: Aggregate gene abundances to estimate functional pathway abundances using integrated KEGG, CAZyme, and antibiotic resistance gene annotations [24]. Identify specific functional modules including Gut Brain Modules, Gut Metabolic Modules, and KEGG modules [24].
- Strain-level Analysis: Track single nucleotide variants in signature genes to monitor strain dissemination and dynamics [24].
The following workflow diagram illustrates the integrated experimental and computational approach for studying microbiome-drug interactions:
Workflow for Microbiome-Drug Interaction Studies
Validation and Functional Assays
Materials:
- Anaerobic chamber for bacterial cultivation
- Gnotobiotic mouse models
- Targeted metabolomics platforms (e.g., LC-MS/MS for SCFA, bile acids)
- In vitro fermentation systems
Protocol:
- Microbial Cultivation: Isolate specific drug-metabolizing bacteria under anaerobic conditions (2-5% Hâ‚‚, 5-10% COâ‚‚, balance Nâ‚‚) using appropriate media [68].
- In Vitro Drug Metabolism Assays: Incubate test drugs with bacterial pure cultures or defined communities in anaerobic buffer systems. Monitor drug depletion and metabolite formation using LC-MS/MS [68] [67].
- Gnotobiotic Mouse Studies: Colonize germ-free mice with defined microbial communities exhibiting specific drug-metabolizing capabilities. Administer test drugs and compare pharmacokinetic parameters to conventional or germ-free controls [68] [67].
- Metabolomic Validation: Perform targeted metabolomics on patient samples to validate microbial metabolites predicted from metagenomic analyses, particularly short-chain fatty acids, bile acids, and tryptophan metabolites [70].
Research Reagent Solutions
Table 3: Essential Research Reagents for Microbiome-Drug Interaction Studies
| Reagent/Catalog | Manufacturer | Function/Application |
|---|---|---|
| MP-soil FastDNA Spin Kit for Soil | MP Biomedicals | High-yield microbial DNA extraction from complex samples |
| NEBNext Microbiome DNA Enrichment Kit | New England Biolabs | Host DNA depletion to increase microbial sequencing depth |
| Illumina DNA Prep Kit | Illumina | Library preparation for shotgun metagenomic sequencing |
| PacBio HiFi SMRTbell Libraries | PacBio | Long-read metagenomic sequencing for improved assembly |
| Meteor2 Software | Open Source | Integrated taxonomic, functional, and strain-level profiling |
| KEGG Database | Kanehisa Laboratories | Functional annotation of microbial metabolic pathways |
| dbCAN3 Database | Carbohydrate-active enzyme annotation | |
| ResFinder/ResFinderFG | Antibiotic resistance gene identification | |
| Anaerobic Chamber | Coy Laboratory | Cultivation of oxygen-sensitive gut microorganisms |
| Germ-Free Mouse Models | Jackson Laboratory | In vivo validation of microbial drug metabolism |
Shotgun metagenomic approaches have fundamentally transformed our understanding of microbiome-drug interactions, revealing the extensive capability of gut microorganisms to directly and indirectly modulate drug metabolism and treatment outcomes. The integration of advanced bioinformatic tools like Meteor2 with multi-omics frameworks and experimental validation provides a powerful platform for deciphering the complex mechanisms underlying pharmacomicrobiomic interactions [24] [70]. As this field advances, key challenges remain, including standardization of analytical protocols, development of reference materials, improved functional annotation of microbial genes, and equitable representation of global populations in microbiome studies [70].
Future research directions should prioritize longitudinal study designs to capture dynamic microbiome-drug interactions, development of personalized microbial community models for predictive toxicology, and clinical translation of pharmacomicrobiomic biomarkers for treatment stratification. Ultimately, integrating microbiome data with pharmacogenomic and clinical variables will enable truly comprehensive precision medicine approaches that optimize therapeutic efficacy while minimizing adverse drug reactions across diverse patient populations [67]. The ongoing development of advanced sequencing technologies, analytical methods, and multi-omic integration frameworks promises to accelerate these discoveries, potentially revolutionizing how we approach drug development and personalized therapy in the coming decade.
Navigating Analytical Challenges: Troubleshooting and Optimization Strategies
In shotgun metagenomic sequencing for microbiome functional analysis, host nucleic acid contamination presents a significant barrier to data quality and interpretation. The overwhelming abundance of host-derived DNA in samples from host-associated environments can obscure microbial signals, reducing sequencing depth and sensitivity for detecting pathogenic or commensal microorganisms [71]. This challenge is particularly acute in low microbial biomass environments, such as the respiratory tract, where host DNA can comprise over 99.9% of total sequenced DNA, effectively drowning out the microbial signal of interest [71]. The development and implementation of robust host depletion strategies are therefore critical prerequisites for accurate profiling of microbial communities and their functional capacities. This application note provides a comprehensive overview of contemporary methodologies for identifying and mitigating host DNA contamination, enabling researchers to generate more reliable and interpretable metagenomic data.
Host DNA Depletion Methodologies
Host DNA depletion methods can be broadly categorized into pre-extraction and post-extraction approaches, each with distinct mechanisms, advantages, and limitations. Pre-extraction methods physically separate or selectively degrade host material prior to DNA extraction, while post-extraction methods enrich for microbial DNA after total nucleic acid extraction.
Table 1: Comparison of Host DNA Depletion Methods for Respiratory Samples
| Method | Mechanism | Host DNA Reduction | Microbial DNA Retention | Key Limitations |
|---|---|---|---|---|
| Saponin Lysis + Nuclease (S_ase) | Lysis of human cells with saponin followed by nuclease digestion | High (to 0.01% of original) | Moderate | Diminishes certain commensals and pathogens |
| HostZERO Kit (K_zym) | Selective lysis of human cells | High (to 0.01% of original) | Moderate to Low | Introduces contamination, alters microbial abundance |
| Filtering + Nuclease (F_ase) | Size-based separation followed by nuclease digestion | Moderate | Moderate (most balanced performance) | Requires optimization for different sample types |
| Nuclease Digestion (R_ase) | Digestion of free DNA without cellular lysis | Low to Moderate | High (31% in BALF) | Less effective for intracellular host DNA |
| Osmotic Lysis + PMA (O_pma) | Hypotonic lysis followed by PMA degradation | Low (2.5-fold increase) | Low | Least effective for increasing microbial reads |
| NEBNext Microbiome Kit | Methylation-based enrichment | Poor for respiratory samples | Variable | Inconsistent performance across sample types |
The efficacy of these methods varies significantly by sample type. In bronchoalveolar lavage fluid (BALF) samples, which typically contain high host DNA content (median 4446.16 ng/mL) and low bacterial load (median 1.28 ng/mL), the HostZERO Microbial DNA Kit (Kzym) demonstrated the best performance in increasing microbial reads (100.3-fold increase), followed by saponin lysis with nuclease digestion (Sase, 55.8-fold increase) [71]. However, these efficiency gains must be balanced against potential biases, as all host depletion methods can introduce contamination, alter microbial abundance, and differentially impact specific bacterial taxa such as Prevotella spp. and Mycoplasma pneumoniae [71].
Experimental Protocols for Host DNA Depletion
Protocol 1: Saponin-Based Host Depletion for Respiratory Samples
This protocol has been optimized for bronchoalveolar lavage fluid (BALF) and oropharyngeal swab (OP) samples [71].
Reagents and Equipment:
- Saponin stock solution (0.025% in PBS)
- DNase I (RNase-free)
- DNase reaction buffer
- Proteinase K
- PBS (phosphate-buffered saline)
- Thermal mixer
- Centrifuge
- Magnetic stand for clean-up
Procedure:
- Sample Preparation: Centrifuge 1-2 mL of BALF at 5,000 × g for 10 minutes. For swabs, elute in 1 mL PBS.
- Human Cell Lysis: Resuspend pellet in 200 μL of 0.025% saponin solution. Vortex thoroughly and incubate at room temperature for 10 minutes.
- Nuclease Digestion: Add 10 μL DNase I and 25 μL 10× DNase reaction buffer. Mix gently and incubate at 37°C for 30 minutes.
- Enzyme Inactivation: Add 20 μL of 50 mM EDTA and heat at 75°C for 10 minutes.
- Microbial Lysis: Add 200 μL of lysis buffer containing proteinase K. Incubate at 56°C for 30 minutes.
- DNA Purification: Purify DNA using magnetic bead-based clean-up according to manufacturer's instructions.
- Quality Assessment: Quantify DNA using fluorometric methods and assess host DNA depletion via qPCR targeting human-specific genes.
Technical Notes: The 0.025% saponin concentration was determined to be optimal through concentration testing. Include negative controls (saline processed similarly) to monitor contamination. Cell-free microbial DNA (approximately 69% in BALF and 80% in OP) will not be captured by this method [71].
Protocol 2: SIFT-seq for Contamination-Resistant Metagenomic Sequencing
Sample-Intrinsic microbial DNA Found by Tagging and sequencing (SIFT-seq) implements a novel approach to wet-lab metagenomics that makes sequencing robust against environmental DNA contamination introduced during sample preparation [72].
Reagents and Equipment:
- Bisulfite conversion kit
- DNA isolation kit
- Library preparation reagents
- Thermal cycler
- DNA quantification system
Procedure:
- Sample Tagging: Add bisulfite conversion reagent directly to 200-500 μL of raw sample (plasma, urine, or other liquid sample). Incubate according to manufacturer's instructions to convert unmethylated cytosines to uracils.
- DNA Extraction: Isolate DNA using standard methods appropriate for the sample type.
- Library Preparation: Prepare sequencing libraries using standard metagenomic protocols.
- Bioinformatic Filtering:
- Remove host-derived sequences via alignment to host reference genome.
- Eliminate sequences containing CG dinucleotides (indicative of methylated host DNA).
- Flag and remove sequences with more than three cytosines or one cytosine-guanine dinucleotide.
- Apply species-level filtering to remove reads originating from C-poor regions in reference genomes.
Technical Notes: SIFT-seq reduces contaminant genera by up to three orders of magnitude and can completely remove common contaminants like Cutibacterium acnes from certain samples [72]. This method is particularly valuable for low-biomass applications such as cell-free DNA sequencing from blood or urine.
Computational Approaches for Contaminant Identification
Beyond wet-lab methods, computational tools provide essential post-sequencing approaches for identifying and removing contaminant sequences. The decontam R package implements two statistical classification methods for distinguishing contaminants from true microbial sequences based on patterns observed in sequencing data [73].
Table 2: Computational Contaminant Identification Methods
| Method | Principle | Requirements | Best Applications |
|---|---|---|---|
| Frequency-Based | Contaminant frequency inversely correlates with sample DNA concentration | DNA quantitation data | Higher biomass samples with variable DNA concentration |
| Prevalence-Based | Contaminants have higher prevalence in negative controls than true samples | Sequenced negative controls | All sample types, especially with consistent controls |
| Combined | Integrates both frequency and prevalence patterns | DNA quantitation + negative controls | Most comprehensive approach |
Implementation with decontam:
- Frequency-Based Method: This approach assumes contaminating DNA is present at relatively uniform levels across samples, while true sample DNA varies with biomass. For each sequence feature, two linear models are fit to log-transformed frequencies: a contaminant model with slope -1 and a non-contaminant model with slope 0. The score statistic is calculated as the tail probability at value R of an F distribution [73].
- Prevalence-Based Method: This method exploits the tendency of contaminants to appear more frequently in negative controls than true samples due to the absence of competing DNA. A chi-square test on the 2×2 presence-absence table in true samples versus negative controls generates a score statistic for classification [73].
Best Practices for Contamination Prevention
Effective management of host DNA contamination begins with rigorous preventive measures throughout the experimental workflow [74].
Sample Collection and Handling
- Decontaminate Sources: Treat equipment, tools, and vessels with 80% ethanol followed by a nucleic acid-degrading solution (e.g., sodium hypochlorite, UV-C exposure).
- Use Personal Protective Equipment (PPE): Wear gloves, masks, and protective clothing to limit contact between samples and contamination sources.
- Employ Single-Use DNA-Free Consumables: Whenever possible, use pre-sterilized collection materials.
- Include Sampling Controls: Process empty collection vessels, exposed swabs, and preservation solutions alongside biological samples.
Laboratory Processing
- Physical Separation: Maintain separate areas for pre- and post-PCR activities.
- Reagent Validation: Test molecular biology reagents for microbial DNA contamination before use.
- Equipment Sterilization: Treat reusable equipment with ethylene oxide gas, which alkylates amine groups in DNA causing mutations and strand breaks that prevent amplification [75].
- Negative Controls: Include extraction blanks and PCR no-template controls in every batch.
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions for Host DNA Depletion
| Reagent/Kit | Function | Application Notes |
|---|---|---|
| Saponin | Detergent that selectively lyses mammalian cells without disrupting bacterial cell walls | Optimal at 0.025% concentration for respiratory samples; cost-effective |
| DNase I | Endonuclease that cleaves free DNA without membrane permeabilization | Effective for degrading host DNA released during lysis steps |
| Propidium Monoazide (PMA) | DNA intercalating dye that penetrates compromised membranes; photoactivatable for DNA cross-linking | Selectively degrades DNA from dead cells; use at 10 μM concentration |
| HostZERO Microbial DNA Kit | Commercial kit for selective depletion of host DNA | High efficiency for respiratory samples; reduces host DNA to 0.01% of original |
| QIAamp DNA Microbiome Kit | Commercial system for enrichment of microbial DNA | Moderate host depletion with good bacterial retention (21% in OP samples) |
| Bisulfite Conversion Reagents | Chemical conversion of unmethylated cytosine to uracil | Foundation of SIFT-seq; tags intrinsic DNA for contamination removal |
| Ethylene Oxide Gas | Alkylating agent for sterilizing consumables | Damages DNA through alkylation of amine groups, preventing amplification |
Workflow Visualization
Effective management of host DNA contamination requires a multifaceted approach spanning careful experimental design, appropriate depletion methodology selection, and rigorous bioinformatic processing. No single method universally addresses all challenges, and optimal approaches must be determined by sample type, biomass, and research objectives. The F_ase (filtering with nuclease digestion) method has demonstrated the most balanced performance for respiratory samples, while emerging technologies like SIFT-seq offer promising avenues for contamination-resistant metagenomic sequencing [71] [72]. By implementing these strategies and maintaining stringent contamination controls throughout the workflow, researchers can significantly enhance the reliability and interpretability of shotgun metagenomic data for microbiome functional analysis.
Shallow shotgun metagenomic sequencing represents a transformative approach for large-scale microbiome studies, offering a superior alternative to 16S rRNA sequencing at a cost substantially lower than deep shotgun metagenomics. This application note examines the technical considerations, experimental protocols, and analytical frameworks for implementing shallow shotgun sequencing to balance budgetary constraints with requirements for taxonomic and functional resolution. We provide validated methodologies and benchmarking data to guide researchers in designing robust microbiome studies for therapeutic development and clinical applications.
Microbiome research has evolved beyond taxonomic cataloging to functional analysis, demanding sequencing strategies that provide genetic resolution while accommodating sample throughput requirements. Shotgun metagenomic sequencing enables comprehensive analysis of microbial communities by randomly sequencing all DNA fragments in a sample, providing access to the full genetic content and functional potential of microbiomes [76]. While deep shotgun sequencing (typically >10 million reads per sample) enables strain-level characterization and single nucleotide variant identification, its cost can be prohibitive for large cohort studies [77] [78].
Shallow shotgun sequencing (SSMS) has emerged as a viable intermediate, generating approximately 0.5-5 million reads per sample at a cost comparable to 16S amplicon sequencing while providing species-level taxonomic resolution and direct functional profiling [77] [79] [78]. This approach is particularly valuable for large-scale biomarker discovery, longitudinal studies, and population-level screening where statistical power requires hundreds to thousands of samples [78].
Comparative Methodologies
Technical Comparison of Sequencing Approaches
Table 1: Comparison of microbiome sequencing methodologies
| Parameter | 16S Amplicon Sequencing | Shallow Shotgun Sequencing | Deep Shotgun Sequencing |
|---|---|---|---|
| Sequencing Cost | Low | Moderate (2-3x 16S) | High (5-10x 16S) |
| Taxonomic Resolution | Genus-level (sometimes species) | Species-level | Species to strain-level |
| Functional Profiling | Predicted (imputed) | Direct measurement | Comprehensive direct measurement |
| Organisms Detected | Bacteria & Archaea (16S); Fungi (ITS) | All domains: bacteria, archaea, eukaryotes, viruses | All domains with higher sensitivity |
| Host DNA Contamination | Low (targeted amplification) | High (requires management) | High (requires management) |
| Technical Variation | Higher | Lower [78] | Lower |
| Ideal Application | Large cohort taxonomic screening | Large studies requiring species-level taxonomy & function | Strain characterization, genetic variation studies |
Quantitative Performance Benchmarks
Table 2: Performance characteristics of shallow shotgun sequencing
| Metric | Shallow Shotgun Performance | Reference |
|---|---|---|
| Recommended Depth | 0.5-5 million reads/sample | [77] [79] |
| Correlation with Deep Sequencing | 97% for species composition; 99% for metagenomic profiles | [77] |
| Species-Level Classification | ~62.5% of reads assigned to species/strain level | [78] |
| Technical Variation | Significantly lower than 16S sequencing | [78] |
| Cost Relative to 16S | Moderately higher (similar when considering information content) | [79] |
| Minimum DNA Input | 2 ng for library preparation | [79] |
Experimental Protocol
Sample Preparation and DNA Extraction
Principle: High-quality, inhibitor-free DNA extraction is critical for all downstream analyses. The protocol must be optimized for sample type to maximize microbial representation while minimizing host DNA contamination.
Validated Protocol:
- Extraction Kit: Qiagen MagAttract PowerSoil DNA KF Kit (optimized for Thermofisher KingFisher robot) [79]
- Mechanism: Magnetic beads specifically capture DNA while excluding organic inhibitors
- Quality Assessment: Fluorometric quantification and fragment analysis
- Sample Types Validated: Stool, soil, water; lower biomass samples may require protocol modifications
Sample Type Considerations:
- High host DNA samples (skin, tissue, blood): Host depletion may be necessary; 16S may be preferred [80]
- Low biomass samples: Increased sequencing depth may be required despite shallow approach
- Fecal samples: Ideal for SSMS with high microbial biomass [79]
Library Preparation and Sequencing
Principle: Library preparation converts extracted DNA into sequencing-ready libraries while maintaining representation of community diversity.
Validated Protocol:
- Library Prep Kit: Illumina Nextera Flex DNA library prep kit [79]
- Alternative Options: seqWell plexWell 96 for higher throughput [81]
- Sequencing Platform: Illumina NextSeq for balanced throughput and cost [79]
- Read Configuration: Paired-end sequencing (2×150 bp recommended)
- Multiplexing: 96-384 samples per lane depending on desired depth
Method Selection Considerations:
- Miniaturization approaches reduce costs but may impact workflow efficiency [81]
- Automation-friendly formats (e.g., plexWell 96) significantly reduce hands-on time for large studies [81]
- Cost-effective library prep methods perform comparably to standard methods across diverse sample types [81]
Bioinformatic Analysis
Principle: Appropriate bioinformatic processing is essential to convert raw sequencing data into biologically meaningful information.
Standard Workflow:
- Quality Control: FastQC for read quality assessment
- Adapter Trimming: Trimmomatic or Cutadapt
- Host DNA Depletion: Alignment to host genome (e.g., hg38) and removal of matching reads
- Taxonomic Profiling: k-mer based alignment to curated reference databases (RefSeq) [79]
- Functional Annotation: Mapping to KEGG, SEED, or COG databases [79]
Deliverables:
- Processed, high-quality FASTQ files
- Taxonomic abundance tables (species and genus level)
- Functional profiles (KEGG orthology groups, modules, and pathways)
- Alpha and beta diversity metrics
- Differential abundance analysis
Workflow Integration
Diagram 1: Experimental workflow for shallow shotgun metagenomic sequencing - This diagram illustrates the complete pipeline from sample collection to data interpretation, highlighting critical decision points for sample-specific processing.
Research Reagent Solutions
Table 3: Essential research reagents and materials for shallow shotgun metagenomic sequencing
| Component | Product Examples | Function & Application Notes |
|---|---|---|
| DNA Extraction | Qiagen MagAttract PowerSoil DNA KF Kit | Magnetic bead-based purification; optimal yield/quality balance for diverse sample types [79] |
| Library Preparation | Illumina Nextera Flex DNA Library Kit | Tagmentation-based fragmentation and tagging; efficient for low-input samples [79] |
| High-Throughput Library Prep | seqWell plexWell 96 | 96-well library prep; significantly reduces hands-on time and cost [81] |
| Alternative Library Prep | QIASeq FX DNA, NextFlex Rapid DNA-Seq | Cost-effective alternatives with varying performance characteristics [81] |
| Sequencing Platform | Illumina NextSeq 550/2000 | Ideal balance of throughput and cost for shallow sequencing applications [79] |
| Quality Control | Agilent Bioanalyzer/TapeStation, Qubit Fluorometer | Essential for DNA quantification and quality assessment prior to library prep |
Application Scenarios and Decision Framework
Study Design Considerations
Ideal Applications for Shallow Shotgun Sequencing:
- Large cohort studies (>100 samples) requiring species-level resolution
- Longitudinal designs with frequent sampling [78]
- Biomarker discovery where functional potential adds value beyond taxonomy
- Population screening studies balancing budget and information content
When to Consider Alternative Approaches:
- Strain-level characterization requires deep sequencing (>20 million reads) [77]
- Low-abundance taxa detection (<0.1% abundance) necessitates deeper sequencing [77]
- Single nucleotide variant (SNV) identification requires ultra-deep sequencing [77]
- Studies with extreme budget constraints may begin with 16S sequencing
Sample Type-Specific Recommendations
High-Success Sample Types:
- Fecal samples: Ideal for SSMS with high microbial biomass and diversity [79]
- Soil samples: High diversity but may require deeper sequencing for rare taxa
- Environmental water: Variable success depending on biomass concentration
Challenging Sample Types:
- Skin swabs: Often contain >90% host DNA; may require deeper sequencing or 16S [77]
- Tissue biopsies: High host contamination; specialized host depletion needed
- Blood samples: Low microbial biomass; may not be suitable for SSMS
Shallow shotgun metagenomic sequencing represents an optimal balance between cost and resolution for large-scale microbiome studies. By providing species-level taxonomic classification and direct functional profiling at a cost substantially lower than deep shotgun sequencing, SSMS enables researchers to design appropriately powered studies without sacrificing genetic resolution. The protocols and considerations outlined herein provide a framework for implementing this approach across diverse research applications, from therapeutic development to population health studies. As sequencing costs continue to decline and reference databases expand, SSMS is positioned to become the standard approach for cohort-scale microbiome functional analysis.
Assembly Challenges in High-Diversity Communities and Solutions
Shotgun metagenomic sequencing has revolutionized microbial ecology by enabling the comprehensive study of microbial communities directly from their environment, without the need for cultivation [23] [82]. However, the assembly of these sequences remains a significant computational challenge, particularly in high-diversity environments where numerous closely related strains coexist [83]. These communities present obstacles such as sequence redundancy, repetitive regions, and uneven microbial abundances, which can lead to fragmented assemblies and misassembled genomes [82] [83]. Overcoming these challenges is crucial for accurate taxonomic profiling, functional characterization, and the reconstruction of metagenome-assembled genomes (MAGs), which are essential for understanding microbial roles in health, disease, and ecosystem functioning [57] [82]. This application note details these challenges and presents standardized protocols and solutions for effective metagenomic assembly in complex communities.
Key Challenges in Metagenomic Assembly
The process of reconstructing complete genomes from complex microbial communities is fraught with technical difficulties that compromise assembly quality and completeness.
- High Microbial Diversity and Strain Heterogeneity: Environments like soil and the human gut contain thousands of microbial species, many with numerous closely related strains. This diversity leads to shared genomic regions that confound assembly algorithms, resulting in fragmented contigs rather than complete genomes [82] [83].
- Uneven Abundance and Low Coverage of Species: Microbial communities often have a dominant minority of species and a long tail of low-abundance organisms. Sequencing depth may be insufficient to cover these rare species, making it impossible to assemble their genomes fully. This phenomenon contributes to the "microbial dark matter" that remains uncharacterized [82].
- Repetitive and Mobile Genetic Elements: Repetitive sequences, including transposons and insertion sequences, are common in microbial genomes. These regions are challenging for assemblers to resolve, frequently causing assembly breaks and misassemblies [83].
- Limitations in Sequencing Technologies: While short-read sequencing (e.g., Illumina) offers high accuracy, it produces reads that are too short to span repetitive regions. Long-read technologies (e.g., Oxford Nanopore, PacBio) can address this but have higher error rates and lower throughput, complicating the assembly process [23].
Comparative Analysis of Assembly Solutions
The table below summarizes the primary strategies developed to address assembly challenges, along with their performance characteristics.
Table 1: Solutions for Metagenomic Assembly Challenges in High-Diversity Communities
| Solution Approach | Key Methodology | Reported Performance and Advantages | Associated Tools / Examples |
|---|---|---|---|
| Co-assembly | Pooling and jointly assembling sequencing reads from multiple related samples [83]. | Increases effective sequencing depth, improves genome fraction (e.g., 4.94% vs. 4.83% in one study), reduces duplication ratio (1.09 vs. 1.23), and yields longer contigs [83]. | metaSPAdes, MEGAHIT |
| Specialized Gene Catalogues | Using environment-specific reference databases of microbial genes for read mapping and quantification, bypassing de novo assembly [57]. | Improves sensitivity for low-abundance species (≥45% increase) and functional abundance accuracy (≥35% improvement). Enables fast profiling (e.g., 2.3 min for taxonomy) [57]. | Meteor2 |
| Hybrid Assembly | Integrating both short-read (high accuracy) and long-read (long span) sequencing data [23] [83]. | Resolves repetitive regions and improves contiguity of assemblies, leading to more complete MAGs. | Unspecified in results |
| Advanced Binning Techniques | Grouping assembled contigs into genome bins using sequence composition (GC content, k-mers) and abundance profiles across samples [82]. | Essential for reconstructing genomes from uncultured organisms, dramatically expanding the known microbial tree of life (MAGs represent ~48% of bacterial diversity) [82]. | MSPminer |
Detailed Experimental Protocols
Protocol 1: Metagenomic Co-assembly for Enhanced Gene Recovery
This protocol is designed to improve assembly quality in low-biomass or high-diversity samples, such as airborne microbiomes, by leveraging co-assembly [83].
I. Sample Preparation and Sequencing
- Sample Collection: Collect multiple metagenomic samples from a similar environmental niche or experimental condition. For air samples, use filters and collect during defined events (e.g., dust storms) [83].
- DNA Extraction: Use a kit designed for low-biomass samples (e.g., DNeasy PowerSoil Kit). Elute DNA in a low-EDTA TE buffer to preserve integrity.
- Library Preparation and Sequencing: Prepare libraries with a dual-indexing strategy (e.g., Illumina Nextera XT). Sequence on a platform such as Illumina NovaSeq to a minimum depth of 4-5 million paired-end reads per sample after quality control [83].
II. Bioinformatics Processing
- Quality Control and Trimming: Process raw reads (e.g., FASTQ files) with Trimmomatic [23] to remove adapters and low-quality bases.
- Co-assembly: Pool quality-filtered reads from all samples in a subgroup and perform co-assembly.
- Assembly Quality Assessment: Evaluate the co-assembly using QUAST with a custom reference genome set representative of the sample environment [83]. Key metrics include genome fraction, duplication ratio, and number of misassemblies.
Protocol 2: Taxonomic and Functional Profiling Using Specialized Gene Catalogues
This protocol uses the Meteor2 pipeline for comprehensive taxonomic, functional, and strain-level profiling (TFSP) without assembly, ideal for well-characterized environments like the human gut [57].
I. Data Preprocessing and Mapping
- Read Trimming and Host Removal: Trim reads to 80 nucleotides and remove host-derived sequences using a tool like Bowtie2 against the host genome.
- Read Mapping to a Gene Catalogue: Map non-host reads to a specialized microbial gene catalogue (e.g., the human gut catalogue in Meteor2) using Bowtie2 with stringent parameters.
II. Profiling and Abundance Estimation
- Gene and Species Abundance Estimation: Use Meteor2's built-in functions to generate abundance profiles from the mapping file (BAM). The default "shared" counting mode proportionally distributes multi-mapping reads.
- Functional Annotation: Annotate the abundance profile using Meteor2's integrated databases (KEGG Orthology, CAZymes, Antibiotic-Resistant Genes).
- Strain-Level Tracking: Execute the strain-level analysis module to track single nucleotide variants (SNVs) in signature genes.
Workflow Visualization
The following diagram illustrates the logical workflow and decision points for the two primary protocols described above, helping researchers select the appropriate path for their specific research goals.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful execution of the protocols requires specific reagents, databases, and software tools. The table below lists the essential components.
Table 2: Key Research Reagents and Materials for Metagenomic Assembly and Profiling
| Category | Item / Tool | Function / Application | Example / Specification |
|---|---|---|---|
| Wet-Lab Reagents | DNA Extraction Kit | Isolation of high-molecular-weight DNA from complex samples. | Kits for low-biomass (e.g., for air filters [83]) or high-inhibitor (e.g., soil) samples. |
| Nucleic Acid Preservation Buffer | Stabilizes microbial community DNA/RNA between collection and processing. | RNAlater, OMNIgene.GUT [82]. | |
| Library Prep Kit | Prepares sequencing libraries from metagenomic DNA. | Illumina Nextera XT, Illumina DNA Prep. | |
| Bioinformatics Tools | Quality Control Tool | Assesses read quality and filters/trims low-quality data. | Trimmomatic [23], FASTQC [23]. |
| Assembly Software | Reconstructs longer sequences (contigs) from short reads. | metaSPAdes, MEGAHIT [84]. | |
| Binning Software | Groups contigs into putative genomes (MAGs). | MSPminer [57]. | |
| Profiling Pipeline | Performs taxonomic/functional profiling from reads. | Meteor2 [57]. | |
| Reference Databases | Microbial Gene Catalogue | Environment-specific reference for gene-based profiling. | Meteor2 catalogues (human gut, oral, etc.) [57]. |
| Functional Database | Annotates genes with functional terms. | KEGG [57], dbCAN3 (CAZymes) [57]. | |
| Taxonomic Database | Provides a reference for taxonomic classification. | Genome Taxonomy Database (GTDB) [57]. |
Shotgun metagenomic sequencing has revolutionized microbiome research by enabling comprehensive analysis of microbial community composition and functional potential directly from environmental samples [2] [1]. Unlike targeted amplicon sequencing, this approach sequences all genomic DNA within a sample, providing insights into bacteria, archaea, fungi, viruses, and their collective genetic capabilities [2]. The computational challenges associated with processing, analyzing, and managing the vast datasets generated by shotgun metagenomics present significant hurdles for researchers [85]. This application note provides detailed protocols and frameworks for managing computational resources and data throughout the metagenomic analysis workflow, with specific consideration for large-scale studies in microbiome functional analysis research.
Experimental Design and Sample Processing
Pre-sequencing Considerations
Effective metagenomic analysis begins with careful experimental design and sample processing. A clearly defined research goal is essential to guide computational strategy and resource allocation [86]. Key considerations include defining the ecological aspect of interest, formulating specific research questions, reviewing existing literature for experimental controls, and identifying potential comparative analyses [86].
Sample collection protocols must prioritize sterility, immediate freezing at appropriate temperatures (-20°C or -80°C), and minimization of freeze-thaw cycles to preserve microbial community integrity [2]. For host-associated microbiomes, fractionation or selective lysis may be necessary to minimize host DNA contamination, which can overwhelm microbial signals in sequencing data [37]. DNA extraction method selection significantly influences microbial community representation, with direct lysis methods typically providing higher yields but potential biases in diversity representation [37].
Sequencing Technology Selection
The choice between short-read and long-read sequencing technologies significantly impacts downstream computational requirements and analytical possibilities [86].
Table 1: Comparison of Sequencing Technologies for Metagenomics
| Platform Type | Read Length | Advantages | Limitations | Best Applications |
|---|---|---|---|---|
| Short-read (Illumina) | 150-300 bp | Cost-effective, high accuracy, high throughput | Difficulties with repetitive regions and strain differentiation | High-coverage surveys, functional profiling [86] |
| Long-read (PacBio, Nanopore) | 10,000-50,000+ bp | Resolves repetitive regions, improves genome completeness | Higher cost per base, requires high-quality DNA | Metagenome-assembled genomes (MAGs), complex regions [86] |
For large-scale studies, shallow shotgun sequencing provides a cost-effective alternative to full-depth sequencing while maintaining higher discriminatory power than 16S sequencing [1]. Sequencing depth should be optimized based on research objectives, with deeper coverage required for strain-level differentiation and variant calling [2].
Computational Workflow and Analysis Strategies
Shotgun metagenomic data analysis typically follows two primary approaches: read-based (assembly-free) and assembly-based analyses [86]. The choice between these methods depends on research objectives, sample complexity, and computational resources.
Read-based analysis involves directly comparing sequencing reads against reference databases without prior assembly. This approach is computationally efficient and retrieves more functional predictions but may overpredict genes and depends heavily on reference database quality [86]. It is particularly suitable for well-characterized environments where reference databases adequately represent community diversity.
Assembly-based analysis involves reconstructing longer contiguous sequences (contigs) from short reads before analysis. This approach enables identification of novel taxa and genes, provides genomic context, and improves taxonomic classification but demands substantial computational resources and is challenged by highly complex communities [86].
For large-scale studies, hybrid approaches such as implemented in MetaflowX offer resource-efficient solutions, completing analyses up to 14-fold faster with 38% less disk usage while recovering more high-quality MAGs compared to conventional workflows [87].
Bioinformatic Workflow
The following diagram illustrates the comprehensive computational workflow for shotgun metagenomic analysis:
Workflow Diagram 1: Comprehensive Metagenomic Analysis Pipeline
Detailed Methodological Protocols
Quality Control and Preprocessing
Protocol: Raw Read Processing
- Input: Paired-end or single-end FASTQ files from sequencing facility
- Tools: Trimmomatic, FastQC, Cutadapt [88]
- Steps:
- Assess read quality using FastQC (per-base sequence quality, adapter content, GC distribution)
- Remove adapter sequences and low-quality bases using Trimmomatic
- Filter reads below quality threshold (typically Q20)
- Remove host-derived reads (if applicable) using alignment to host genome
- Quality Metrics: Minimum 70% of reads passing quality filters, Phred score ≥30 for retained bases
Taxonomic Profiling
Protocol: Read-based Taxonomic Classification
- Input: Quality-controlled reads
- Tools: Kraken2, MetaPhlAn [2]
- Steps:
- Pre-built reference database selection (e.g., Standard Kraken2 database or custom-built)
- Classification of reads against database using k-mer based alignment
- Generation of abundance profiles for detected taxa
- Cross-comparison across samples for differential abundance analysis
- Output: Relative abundance tables, phylogenetic profiles, diversity metrics
Metagenome Assembly and Binning
Protocol: Contig Assembly and MAG Generation
- Input: Quality-controlled reads
- Tools: MEGAHIT, MetaSPAdes, MaxBin2, MetaBAT2 [87]
- Steps:
- De novo assembly using appropriate assembler based on data characteristics
- Quality assessment of assembled contigs (N50, length distribution)
- Binning of contigs into putative genomes using composition and abundance information
- MAG refinement using tools like DAS Tool to obtain high-quality genomes
- Quality assessment using CheckM for completeness and contamination estimates
- Quality Thresholds: High-quality MAGs: >90% completeness, <5% contamination; Medium-quality: >50% completeness, <10% contamination [87]
Computational Resource Requirements
Hardware Specifications
Large-scale metagenomic analyses demand substantial computational resources. The following table outlines recommended specifications for different study scales:
Table 2: Computational Resource Requirements for Metagenomic Studies
| Resource Type | Small-scale (10-50 samples) | Medium-scale (50-200 samples) | Large-scale (200+ samples) |
|---|---|---|---|
| CPU Cores | 16-32 cores | 32-64 cores | 64-128+ cores |
| RAM | 64-128 GB | 128-512 GB | 512 GB-2 TB |
| Storage | 1-2 TB (SSD recommended) | 5-10 TB (mixed SSD/HDD) | 20+ TB (with high-speed I/O) |
| Network | Standard 1 GbE | 10 GbE preferred | 25/100 GbE for distributed systems |
| Approximate Processing Time | 2-5 days | 1-2 weeks | 3-6 weeks |
Assembly-based analyses typically require 5-10× more resources than read-based approaches [86]. Recent workflows like MetaflowX demonstrate optimizations reducing disk usage by 38% and completion times by up to 14-fold through efficient resource utilization [87].
Research Reagent Solutions
Table 3: Essential Computational Tools for Metagenomic Analysis
| Tool Category | Specific Tools | Primary Function | Resource Intensity |
|---|---|---|---|
| Quality Control | FastQC, Trimmomatic, Cutadapt | Assess and improve read quality | Low |
| Taxonomic Profiling | Kraken2, MetaPhlAn | Classify reads taxonomically | Medium |
| Assembly | MEGAHIT, MetaSPAdes | Construct contigs from reads | High |
| Binning | MetaBAT2, MaxBin2 | Group contigs into MAGs | High |
| Functional Annotation | HUMAnN2, PROKKA | Identify functional genes and pathways | Medium-High |
| Diversity Analysis | QIIME 2, Phyloseq | Calculate ecological diversity metrics | Low-Medium |
Data Management and Sharing
Data Management Plans
Comprehensive data management is crucial for metagenomic studies. The National Microbiome Data Collaborative (NMDC) recommends developing detailed Data Management Plans (DMPs) that address the entire data lifecycle [89]. Effective DMPs should include:
- Sample and Data Types: Description of data sets, including size, volume, and generation methods
- Data Standards and Formats: Adherence to community standards (GSC MIxS for metagenomics)
- Roles and Responsibilities: Clear assignment of data management tasks within the team
- Data Dissemination and Archiving: Plans for data release timing and target audiences
- Policies for Data Sharing: Compliance with funder requirements for public access
- Data and Sample Preservation: Long-term preservation strategy and budget allocation [89]
Metadata Standards and Repositories
Adherence to standardized metadata schemas ensures data interoperability and reusability. The Genomic Standards Consortium (GSC) Minimum Information about any (x) Sequence (MIxS) standards provide the foundational framework for metagenomic metadata [89]. The following diagram illustrates the data management and sharing workflow:
Workflow Diagram 2: Data Management and Sharing Lifecycle
Table 4: Metadata Standards and Repositories for Microbiome Data
| Data Type | Metadata Standard | Primary Repository | Additional Repositories |
|---|---|---|---|
| Metagenomics | GSC MIxS | SRA (Sequence Read Archive) | ENA, DDBJ [89] |
| Metatranscriptomics | GSC MIxS | Gene Expression Omnibus | ENA, DDBJ [89] |
| Metaproteomics | Proteomics Standards Initiative | PRIDE | ProteomeXchange [89] |
| Metabolomics | Metabolomics Standards Initiative | Metabolomics Workbench | MetaboLights [89] |
FAIR Data Principles Implementation
Making data Findable, Accessible, Interoperable, and Reusable (FAIR) requires strategic planning throughout the research lifecycle [89]. Key implementation strategies include:
- Findability: Assign persistent identifiers (DOIs), rich metadata, and index in searchable resources
- Accessibility: Use standard communication protocols and ensure metadata remains available even if data is restricted
- Interoperability: Use formal knowledge representation languages and qualified references to other metadata
- Reusability: Provide accurate data provenance and meet domain-relevant community standards
Machine-readable data management plans increase the likelihood of data recognition and citation by making data easily locatable and reusable [89].
Effective management of computational resources and data represents a critical success factor for large-scale metagenomic studies. Strategic experimental design, appropriate sequencing technology selection, and thoughtful computational workflow implementation significantly impact research outcomes. The protocols and frameworks presented here provide a foundation for conducting robust metagenomic analyses while ensuring data reproducibility, interoperability, and long-term utility. As the field continues to evolve with emerging technologies like long-read sequencing and machine learning applications, maintaining adherence to community standards and FAIR data principles will remain essential for advancing microbiome science and enabling cross-study comparisons that drive fundamental discoveries in microbial ecology and function.
Optimizing Binning Strategies for Improved Genome Reconstruction from Complex Samples
Shotgun metagenomic sequencing has revolutionized microbiome functional analysis by enabling researchers to reconstruct genomes from complex microbial communities without the need for cultivation. A critical step in this process is metagenomic binning, the computational process of grouping assembled genomic fragments (contigs) into metagenome-assembled genomes (MAGs) that represent individual microbial populations [90] [91]. The quality of binning directly impacts downstream analyses, including functional annotation, comparative genomics, and the identification of biosynthetic gene clusters.
However, optimizing binning strategies remains challenging due to several factors: the immense complexity of microbial communities, limitations in sequencing technologies, and the computational complexity of binning algorithms [90]. This Application Note provides a comprehensive framework for optimizing binning strategies to recover high-quality MAGs from complex samples, with a focus on practical implementation for researchers in microbiome science and drug development.
Binning Performance Across Data Types and Strategies
The performance of binning tools varies significantly across different sequencing technologies and binning modes. A comprehensive benchmark of 13 binning tools revealed that multi-sample binning consistently outperforms other approaches across short-read, long-read, and hybrid data types [92].
Table 1: Performance of Binning Modes Across Sequencing Technologies
| Binning Mode | Data Type | MQ MAGs†| NC MAGs‡ | HQ MAGs§ | Key Applications |
|---|---|---|---|---|---|
| Multi-sample | Short-read (30 samples) | 1101 | 306 | 62 | Large cohort studies with deep sequencing |
| Single-sample | Short-read (30 samples) | 550 | 104 | 34 | Limited sample availability |
| Multi-sample | Long-read (PacBio HiFi) | 1196 | 191 | 163 | Complex regions with repeats |
| Single-sample | Long-read (PacBio HiFi) | 796 | 123 | 104 | Single-sample long-read projects |
| Multi-sample | Hybrid | 827 | 178 | 83 | Maximizing completeness & accuracy |
| Co-assembly | Short-read | Lowest recovery | Lowest recovery | Lowest recovery | Not recommended |
†MQ MAGs: "moderate or higher" quality (completeness >50%, contamination <10%); ‡NC MAGs: near-complete (completeness >90%, contamination <5%); §HQ MAGs: high-quality (completeness >90%, contamination <5%, contains rRNA genes & tRNAs) [92]
Multi-sample binning demonstrated remarkable improvements, recovering 100% more MQ MAGs, 194% more NC MAGs, and 82% more HQ MAGs compared to single-sample binning in marine datasets with 30 metagenomic next-generation sequencing (mNGS) samples [92]. This approach leverages co-abundance patterns across multiple samples to improve bin separation and accuracy.
For long-read data, multi-sample binning showed substantial improvements with larger sample numbers, recovering 50% more MQ MAGs, 55% more NC MAGs, and 57% more HQ MAGs in the marine dataset [92]. The relatively lower sequencing depth in third-generation sequencing makes multi-sample approaches particularly valuable for these data types.
Advanced Binning Tools and Algorithms
Stand-alone Binners and Their Performance
Table 2: High-Performance Binning Tools for Different Data-Binning Combinations
| Tool | Key Algorithm | Optimal Data-Binning Combination | Performance Notes |
|---|---|---|---|
| COMEBin | Contrastive learning with data augmentation | Ranked 1st in 4 combinations | Excellent with multi-sample & hybrid data |
| MetaBinner | Ensemble "partial seed" k-means | Ranked 1st in 2 combinations | Strong multi-tool consensus approach |
| Binny | Iterative HDBSCAN clustering | Ranked 1st in short_read co-assembly | Effective for nonlinear dimensionality reduction |
| VAMB | Variational autoencoders | Efficient binner with good scalability | Deep learning approach for latent representations |
| MetaBAT 2 | Tetranucleotide frequency + coverage | Efficient binner with excellent scalability | Established, reliable performer |
| MetaDecoder | Dirichlet process Gaussian mixture model | Efficient binner | Handles complex distributions well |
Binning Refinement Tools
Bin-refinement tools combine outputs from multiple binning algorithms to reconstruct higher-quality MAGs:
- MetaWRAP: Demonstrates the best overall performance in recovering MQ, NC, and HQ MAGs through bin refinement [92]
- MAGScoT: Achieves comparable performance to MetaWRAP with excellent scalability [92]
- BinSPreader: Protocol available for refining metagenomic binning to improve MAG quality [93]
Experimental Protocols for Optimized Binning
Sample Preparation and Sequencing Strategies
Protocol 4.1.1: Sample Collection and Preservation for Optimal Binning
- Flash Freezing: Collect sample in sterile container and immediately store in dry ice or -80°C conditions [94]
- Preservation Media: Transfer material to microbiome preservation device with nucleic acid-stabilizing buffer [94]
- Biomass Concentration: For complex samples like soil, maximize biomass concentration through centrifugation or filtration to enable deeper sequencing [91]
Protocol 4.1.2: DNA Extraction for Comprehensive Genome Recovery
- Dual Lysis Method: Combine chemical and physical lysis to ensure recovery of both gram-positive and gram-negative organisms [94]
- Quality Control: Assess DNA quantity and quality using fluorometry and fragment analysis
- Library Preparation: Choose between amplicon, shotgun, or hybrid approaches based on research goals [94]
Multi-Sample Binning Protocol
Protocol 4.2.1: Implementation of Multi-Sample Binning
- Sample Requirements: Sequence a minimum of 15-30 samples per cohort for optimal multi-sample binning performance [92]
- Assembly Strategy: Perform individual assembly of each sample using MEGAHIT (short-read) or metaSPAdes with k-mer range 21,41,61,81,99 for complex samples [91]
- Coverage Profiling: Calculate coverage information across all samples for each contig
- Feature Integration: Combine tetranucleotide frequency and coverage profiles using tools like COMEBin or MetaBinner [92]
- Quality Assessment: Evaluate MAG quality using CheckM2 with thresholds of >90% completeness and <5% contamination for near-complete MAGs [92]
Hi-C Binning Protocol for Single Samples
Protocol 4.3.1: Hi-C Assisted Binning for Complex Samples
- Sample Cross-Linking: Resuspend sample in 1% formaldehyde and incubate at room temperature for 20 minutes with periodic mixing [91]
- Quenching: Add glycine powder (1g/100ml) to quench cross-linking and incubate for 15 minutes [91]
- Library Preparation: Use commercial Hi-C kits (e.g., ProxiMeta Hi-C Microbiome Kit) following manufacturer protocols [91]
- Data Integration: Combine Hi-C data with traditional shotgun sequencing using dedicated pipelines
- Mobile Genetic Element Assignment: Leverage Hi-C proximity ligation data to link mobile genetic elements to their host genomes [91]
Coverage-Based Binning Optimization
Protocol 4.4.1: Coverage Analysis with micov Tool
- Input Preparation: Process Sequence Alignment/Map (SAM) files to generate per-sample, per-genome coverage intervals [95]
- Cumulative Coverage Analysis: Plot cumulative coverage breadth across sample groups to identify differentially covered regions
- Bin Optimization: Use coverage patterns to refine bin boundaries and identify chimeric contigs
- Strain Differentiation: Identify strain-specific genomic regions through differential coverage patterns across sample groups [95]
Workflow Visualization
Diagram 1: Comprehensive Metagenomic Binning Workflow
The Scientist's Toolkit: Essential Research Reagents and Computational Tools
Table 3: Essential Research Reagent Solutions for Metagenomic Binning
| Category | Item/Software | Function/Application | Key Features |
|---|---|---|---|
| Wet Lab Reagents | Microbiome Preservation Media | Stabilizes nucleic acids during sample storage | Prevents microbiome shifts post-collection [94] |
| DNA Extraction Kits (e.g., DNeasy PowerWater) | Comprehensive DNA isolation from complex samples | Dual chemical/physical lysis for gram-positive/negative bacteria [94] [91] | |
| ProxiMeta Hi-C Microbiome Kit | Proximity ligation for chromatin conformation capture | Enables Hi-C binning from single samples [91] | |
| Formaldehyde (1%) | Cross-linking for Hi-C library preparation | Preserves physical DNA interactions within cells [91] | |
| Computational Tools | COMEBin | Contrastive learning-based binning | Top performer in multiple data-binning combinations [92] |
| MetaBinner | Ensemble binning algorithm | Ranked 1st in two data-binning combinations [92] | |
| CheckM2 | MAG quality assessment | Evaluates completeness and contamination [92] | |
| micov | Coverage breadth analysis | Identifies differential coverage regions [95] | |
| MetaWRAP | Bin refinement | Best overall performance in MAG refinement [92] | |
| MEGAHIT | Metagenomic assembly | Efficient assembly of complex samples [91] |
Applications in Functional Analysis and Drug Development
Optimized binning strategies enable critical applications in microbiome functional analysis and therapeutic development:
Antibiotic Resistance Gene Host Identification
Multi-sample binning identifies 30% more potential antibiotic resistance gene (ARG) hosts compared to single-sample binning in short-read data, enabling more comprehensive resistance profiling [92]. This capability is crucial for understanding resistance dissemination in clinical and environmental settings.
Biosynthetic Gene Cluster Discovery
Multi-sample binning recovers 54% more potential biosynthetic gene clusters (BGCs) from near-complete strains in short-read data compared to single-sample approaches [92]. This enhanced discovery rate accelerates natural product identification for drug development.
Strain-Level Functional Analysis
Coverage-based tools like micov enable strain-level functional analysis by identifying genomic regions with differential coverage across sample groups [95]. For example, micov identified a genomic region in Prevotella copri that had a stronger effect on overall microbiome composition than the host's country of origin [95].
Optimizing binning strategies through multi-sample approaches, appropriate tool selection, and advanced techniques like Hi-C binning significantly improves genome reconstruction from complex metagenomic samples. The protocols and applications outlined in this document provide researchers with a comprehensive framework for implementing these strategies in microbiome functional analysis and drug development pipelines. As sequencing technologies continue to evolve and computational methods become more sophisticated, these optimized binning approaches will enable deeper insights into microbial community structure and function, accelerating discoveries in human health and disease.
Ensuring Reliability: Validation Techniques and Comparative Method Analysis
In microbiome research, shotgun metagenomic sequencing has become a cornerstone technique for characterizing the genetic potential of microbial communities [7]. It allows researchers to determine "which microbes are present in the community and what they might be doing" by sequencing all DNA content in a sample [7]. However, a significant limitation persists: the presence of genes in metagenomic data only indicates functional potential, not actual activity. This distinction creates a critical validation gap in connecting genetic capacity to real-time microbial function.
Metatranscriptomic analysis addresses this limitation by sequencing RNA molecules, providing a snapshot of genetically active functions and community responses at the time of sampling [96]. The integration of these two methodologies—correlating metagenomic predictions with metatranscriptomic observations—forms a powerful framework for validating functional predictions. This approach moves beyond compositional analysis to reveal the dynamically expressed metabolic pathways that underpin host-microbe interactions [96]. This Application Note provides detailed protocols for designing and executing studies that correlate genetic potential with metatranscriptomic data, enabling robust validation of functional predictions in microbiome research.
Background
Shotgun Metagenomics: Profiling Functional Potential
Shotgun metagenomics involves sequencing all DNA extracted from a community sample, bypassing PCR amplification to provide a less biased view of community structure and function compared to amplicon sequencing [7]. This approach enables researchers to:
- Quantity community structure and diversity
- Assemble novel genomes and identify new taxa
- Determine metabolic pathways encoded in the community [7]
Metagenomic data analysis is computationally complex due to the fragmented nature of the data and the vast diversity of microbial communities. However, reference-based tools have simplified this process by using curated databases of microbial genes and genomes [24].
Metatranscriptomics: Capturing Community Activity
Metatranscriptomics characterizes the RNA content of microbial communities, revealing which genes are being actively transcribed under specific conditions. This approach provides direct insight into microbial gene expression and can identify pathways actively contributing to host health or disease states [96]. A key advantage is the ability to quantify microbial functional pathways as numerical scores, enabling statistical association with host phenotypes such as periodontal disease or nicotine addiction [96].
The Validation Framework
Correlating metagenomic and metatranscriptomic data creates a validation framework where genetic potential (metagenomics) can be confirmed through observed expression (metatranscriptomics). This integration is particularly valuable for:
- Confirming hypothesis generated from metagenomic surveys
- Identifying actively transcribed pathways versus silent genetic capacity
- Understanding microbial community dynamics in response to environmental changes
- Validating therapeutic targets for drug development
Protocol: Integrated Metagenomic-Metatranscriptomic Analysis
This protocol provides a comprehensive methodology for correlating genetic potential with metatranscriptomic data, from sample collection through integrated data analysis.
Sample Collection and Preservation
Proper sample collection and preservation are critical for obtaining high-quality nucleic acids for both DNA and RNA analysis.
Table 1: Sample Collection and Preservation Methods
| Sample Type | DNA Preservation | RNA Preservation | Storage Conditions | Considerations |
|---|---|---|---|---|
| Fecal/Rectal Swab | MP-soil FastDNA Spin Kit [27] | RNA Preservation Buffer (RPB) [96] | -80°C | For rectal swabs, clean with 70% alcohol before collection [27] |
| Saliva | Commercial preservation buffers | RNA Preservation Buffer (RPB) [96] | -80°C | Room temperature stability for up to 28 days with RPB [96] |
| Tissue Biopsy | Flash freezing in liquid N₂ | RNAlater or similar | -80°C | Minimize ischemia time before preservation |
Procedure:
- Collect samples using appropriate sterile techniques to minimize contamination
- For dual DNA/RNA extraction: Split sample immediately after collection into separate preservation systems
- For rectal swabs: Insert sterile swab 4-5 cm, rotate gently, and place in sterile tube [27]
- For saliva: Collect unstimulated saliva into tube with RNA Preservation Buffer [96]
- Record metadata including time of collection, patient phenotype, and clinical parameters
- Store at -80°C until nucleic acid extraction
Nucleic Acid Extraction and Sequencing
Table 2: Nucleic Acid Extraction and Sequencing Parameters
| Step | DNA Protocol | RNA Protocol | Quality Control |
|---|---|---|---|
| Extraction | MP-soil FastDNA Spin Kit [27] | Commercial RNA extraction kits | DNA: NanoDrop, agarose gel [27] |
| Host Depletion | BWA mapping to human genome [27] | Ribosomal RNA depletion kits | RNA: RIN > 7.0 recommended |
| Quality Control | fastp (v 0.23.0) [27] | FastQC or similar | Remove low-quality reads ( |
| Sequencing | Illumina HiSeq 4000, 10-14 Gb [27] | Illumina platform | Minimum 70,000 reads for metatranscriptomics [96] |
Procedure:
- Extract DNA using standardized kits according to manufacturer protocols
- Extract RNA using RNA-specific kits with DNase treatment to remove contaminating DNA
- Assess purity and concentration using spectrophotometry (NanoDrop) and integrity using electrophoresis [27]
- Perform library preparation using Illumina-compatible protocols
- Sequence on appropriate Illumina platform to achieve sufficient depth (≥10 million reads per sample)
Bioinformatic Analysis
The following workflow diagram illustrates the integrated bioinformatic analysis process for correlating metagenomic and metatranscriptomic data:
Metagenomic Analysis Protocol
Tools and Databases:
- Meteor2: For taxonomic, functional, and strain-level profiling [24]
- KEGG database: For functional annotation of orthologous groups [27] [24]
- Custom gene catalogs: Environment-specific references for improved accuracy [24]
Procedure:
- Quality control using fastp (v 0.23.0) to remove adapters and low-quality reads [27]
- Remove host DNA by mapping to human genome using BWA (v 0.7.17) [27]
- Taxonomic profiling using Meteor2 with default parameters (95% identity threshold) [24]
- Functional annotation against KEGG database using Diamond (v 2.0.13) with e-value cutoff of 1e-5 [27]
- Pathway analysis to identify metabolic pathways encoded in the community
Metatranscriptomic Analysis Protocol
Tools and Databases:
- KEGG Orthology (KO): For functional pathway quantification [96]
- Specialized pipelines: For quantifying microbial functional pathways as numerical scores [96]
Procedure:
- Quality control and adapter removal using tools like fastp
- rRNA depletion to enrich for messenger RNA
- Alignment to reference genomes or gene catalogs
- Quantification of gene expression using count-based methods
- Pathway scoring by aggregating expression of genes in functional pathways [96]
Data Integration and Validation
Procedure:
- Normalize data using appropriate methods (e.g., FPKM for metatranscriptomics, depth coverage for metagenomics)
- Compute correlation coefficients between gene abundance (metagenomics) and expression (metatranscriptomics)
- Identify significantly correlated pathways using statistical testing (e.g., Wilcoxon rank-sum test, P < 0.05) [27]
- Visualize relationships using scatter plots, heatmaps, and pathway diagrams
Research Reagent Solutions
Table 3: Essential Research Reagents and Tools
| Category | Product/Software | Specific Function | Application Context |
|---|---|---|---|
| DNA Extraction | MP-soil FastDNA Spin Kit [27] | Microbial DNA extraction from complex samples | Fecal/soil samples with difficult-to-lyse organisms |
| RNA Preservation | RNA Preservation Buffer (RPB) [96] | Maintains RNA integrity at room temperature | Field collections, clinical settings without immediate freezer access |
| Sequencing | Illumina HiSeq 4000 [27] | High-throughput sequencing | Metagenomic and metatranscriptomic library sequencing |
| Taxonomic Profiling | Meteor2 [24] | Taxonomic, functional, strain-level profiling | Integrated analysis using environment-specific gene catalogs |
| Functional Annotation | KEGG Database [27] [24] | Pathway annotation and orthology assignment | Functional interpretation of metagenomic/metatranscriptomic data |
| Quality Control | fastp (v 0.23.0) [27] | Adapter removal and quality filtering | Pre-processing of raw sequencing reads |
| Host DNA Removal | BWA (v 0.7.17) [27] | Maps and removes host-derived reads | Human-associated microbiome studies |
Data Analysis and Interpretation
Effective presentation of quantitative data is essential for interpreting correlation results. The following approaches facilitate clear communication of findings:
Table 4: Statistical Measures for Correlation Analysis
| Metric | Calculation | Interpretation | Application Example |
|---|---|---|---|
| Spearman Correlation | Rank-based correlation coefficient | Measures monotonic relationships | Gene abundance vs. expression correlation |
| Bray-Curtis Dissimilarity | ∑|xi - yi| / ∑(xi + yi) | Quantifies community composition differences | Functional profile comparison between groups [24] |
| Shannon Diversity Index | -∑(pi × ln(pi)) | Measures species diversity considering richness and evenness | α-diversity assessment [27] |
| P-value | Statistical significance testing | Probability of observed results under null hypothesis | Wilcoxon rank-sum test for differential abundance [27] |
Visualization Strategies
The following diagram illustrates the key signaling pathways commonly identified in microbiome studies and their relationship to host health conditions:
Case Study: Acute Pancreatitis Recovery
A recent study on gut microbiota during recovery from acute pancreatitis (AP) demonstrates the application of these correlation principles [27]. The researchers collected rectal swabs from AP patients during both acute and recovery phases, conducting shotgun metagenomic sequencing to analyze gut microbiome composition and function.
Key Findings:
- Microbial diversity remained decreased during recovery compared to healthy controls
- Beneficial bacteria (Bacteroidales) showed increasing tendency in mild AP recovery
- Harmful bacteria (Enterococcus) increased in severe AP recovery phases
- Functional pathways showed opposite trends in recovery phases of different AP severities [27]
This study illustrates how correlating taxonomic composition with functional capacity can inform treatment strategies and prognosis assessment.
Troubleshooting and Optimization
Common Challenges and Solutions
Table 5: Troubleshooting Guide for Integrated Analysis
| Challenge | Potential Cause | Solution | Preventive Measures |
|---|---|---|---|
| Low correlation between datasets | Technical variation, biological timing mismatch | Normalize using different approaches, ensure simultaneous collection | Collect and preserve DNA/RNA from same sample aliquot |
| Poor RNA quality | RNase contamination, improper preservation | Use RNA Preservation Buffer (RPB) [96] | Implement rapid processing, use validated preservation systems |
| High host contamination | Eukaryotic cells in sample | Bioinformatic removal with BWA [27] | Physical separation methods during extraction |
| Limited functional detection | Shallow sequencing depth | Increase sequencing volume (≥10 Gb) [27] | Pilot studies to determine optimal sequencing depth |
| Database limitations | Missing references for novel taxa | Use custom gene catalogs [24] | Environment-specific database construction |
Methodological Considerations
- Sample Collection Timing: Consider diurnal variations in microbial activity when planning collection schedules
- Metadata Collection: Comprehensive clinical and lifestyle data enables more powerful correlations with functional findings [96]
- Statistical Power: Large sample sizes (n > 1000) provide robust detection of associations in heterogeneous populations [96]
- Multiple Testing Correction: Apply false discovery rate corrections when testing multiple hypotheses across many microbial features
The integration of shotgun metagenomic and metatranscriptomic data provides a powerful approach for validating functional predictions in microbiome research. By correlating genetic potential with actual gene expression, researchers can distinguish actively utilized functions from silent genetic capacity, leading to more meaningful biological insights.
The protocols outlined in this Application Note provide a comprehensive framework for designing and executing integrated studies, from proper sample collection through advanced bioinformatic analysis. As recent studies demonstrate [96] [27] [24], this approach can reveal functionally important microbial activities that associate with host health conditions, providing potential targets for therapeutic intervention.
Future methodological developments will likely focus on improving reference databases, enhancing multi-omics integration platforms, and establishing standardized analytical frameworks. By adopting these robust validation approaches, researchers can advance our understanding of microbiome function and its impact on human health and disease.
Within the broader scope of shotgun metagenomic sequencing for microbiome functional analysis, accurate taxonomic profiling serves as a critical foundation. The choice between 16S ribosomal RNA (rRNA) gene amplicon sequencing and whole metagenomic shotgun sequencing represents a fundamental methodological decision that directly impacts the resolution and reliability of downstream biological interpretations [14] [5]. While 16S sequencing targets specific hypervariable regions of this conserved bacterial gene, shotgun sequencing randomly fragments all DNA in a sample, providing a more comprehensive view of the microbial community [14]. This application note systematically benchmarks these competing approaches, providing researchers with structured quantitative comparisons and standardized protocols to guide experimental design in microbiome research, particularly within drug development contexts where accurate species-level identification can inform therapeutic targeting and biomarker discovery.
Performance Comparison: 16S rRNA vs. Shotgun Sequencing
The comparative performance of 16S rRNA and shotgun sequencing methodologies has been quantitatively assessed across multiple studies, focusing on taxonomic resolution, diversity measurements, and detection sensitivity. The table below summarizes key benchmarking findings from recent empirical investigations.
Table 1: Quantitative Comparison of 16S rRNA and Shotgun Sequencing Performance
| Performance Metric | 16S rRNA Sequencing | Shotgun Metagenomics | Supporting Evidence |
|---|---|---|---|
| Taxonomic Resolution | Primarily genus-level, limited species-level identification [5] [97] | Species-level and strain-level discrimination possible [24] [5] | |
| Genus Detection Power | Detects only part of the microbial community [14] [5] | Identifies significantly more genera, particularly low-abundance taxa [14] | Shotgun found 152 significant changes between gut compartments that 16S missed [14] |
| Alpha Diversity | Lower observed diversity [5] | Higher richness estimates [5] | |
| Abundance Correlation | Moderate agreement with shotgun data | Reference standard | Average Pearson correlation: 0.69 at genus level [14] |
| Differential Abundance | Identified 108 significant genera | Identified 256 significant genera | Comparison of caeca vs. crop in chicken model [14] |
| Sensitivity to Sequencing Depth | Maximizes taxa identification with ~50,000 reads [97] | Requires >500,000 reads for stable genus detection [14] | Shotgun samples with <500,000 reads did not reach saturation [14] |
| Functional Profiling | Limited to prediction-based inference [97] | Direct assessment of functional genes and pathways [14] [24] |
The selection of hypervariable regions in 16S sequencing significantly impacts taxonomic resolution. Full-length 16S sequencing (sFL16S) demonstrates improved species-level discrimination compared to partial region sequencing (e.g., V3-V4), with one study reporting 1,041 amplicon sequence variants (ASVs) identified by sFL16S versus 623 ASVs by V3-V4 sequencing [98]. However, even full-length 16S approaches cannot match the comprehensive profiling capacity of shotgun sequencing, which detects a substantially higher number of less abundant but biologically meaningful taxa [14].
Table 2: Impact of 16S rRNA Target Regions on Taxonomic Classification Accuracy
| 16S Region | Key Characteristics | Recommended Applications |
|---|---|---|
| V3-V4 | Standard approach; limited species-level resolution [98] | General microbial profiling when shotgun sequencing is not feasible |
| V1-V3 | Improved taxonomic resolution with concatenation methods [99] | Gut microbiome studies seeking enhanced classification |
| V6-V8 | Superior precision in amplifying gut microbial 16S rRNA genes [99] | Complementary region to V1-V3 for integrated analysis |
| Full-length (V1-V9) | Highest resolution among 16S methods; better species-level identification [98] | Studies requiring maximum 16S resolution without shotgun costs |
Experimental Protocols for Method Comparison
Sample Preparation and Sequencing
DNA Extraction Protocol:
- Stool Sample Processing: Preserve samples in OMR-200 tubes (OMNIgene GUT, DNA Genotek) immediately upon collection, store on ice, and transfer to -80°C within 24 hours [97].
- Extraction Methods: For shotgun sequencing, use NucleoSpin Soil Kit (Macherey-Nagel) following manufacturer's instructions [5]. For 16S sequencing, Dneasy PowerLyzer Powersoil kit (Qiagen) provides optimal results [5].
- DNA Quality Control: Assess integrity through DNA degradation analysis, concentration measurement, and purity verification using Agilent High Sensitivity DNA Bioanalyzer chips [8] [98].
16S rRNA Library Preparation:
- Target Amplification: Amplify the V3-V4 hypervariable region using modified 515FB/806RB primers (515FB: 5'-GTG YCA GCM GCC GCG GTA A-3'; 806RB: 5'-GGA CTA CNV GGG TWT CTA AT-3') [8].
- PCR Conditions: Follow Illumina's standard protocol for 16S amplicon sequencing on MiSeq System with 2×150bp paired-end sequencing [8].
- Library Quality Assessment: Validate library quality using Agilent High Sensitivity DNA Bioanalyzer chips before pooling and sequencing [8].
Shotgun Metagenomic Library Preparation:
- Library Construction: Utilize Nextera XT DNA Library Preparation Kit (Illumina) with Illumina Nextera XT Index v2 Kit A and B following manufacturer's protocols [8].
- Sequencing Parameters: Sequence on Illumina NextSeq500 High Output v2 flowcell producing 2×150bp paired-end reads [8].
- Sequencing Depth: Target minimum 5 million reads per sample for comprehensive taxonomic coverage, with increased depth required for functional analysis [14] [24].
Bioinformatic Analysis Workflows
16S rRNA Data Processing:
- Preprocessing: Use DADA2 (v1.22.0) pipeline for quality filtering, trimming, and error correction [5]. For V3-V4 data, truncate forward reads at 290bp and reverse reads at 230bp with maximum expected errors set to 2 [5].
- Taxonomic Assignment: Assign taxonomy using SILVA 16S rRNA database (v138.1) with minimum confidence threshold of 70% [5] [98].
- Resolution Enhancement: Improve species-level classification by combining SILVA database alignment with k-mer based classification using Kraken2 and Bracken2 against NCBI RefSeq Targeted Loci Project database [5].
Shotgun Metagenomic Data Processing:
- Quality Control and Host Removal: Process raw fastq files with Trim Galore for adapter removal and quality filtering, followed by host sequence removal using KneadData with human genome GRCh38 as reference [8].
- Taxonomic Profiling: For comprehensive taxonomic, functional, and strain-level profiling (TFSP), utilize Meteor2 with environment-specific microbial gene catalogues [24]. Meteor2 employs bowtie2 for read alignment against customized gene catalogues with alignment identity threshold of 95% (98% in fast mode) [24].
- Abundance Estimation: Apply shared counting mode in Meteor2 for gene count calculation, normalizing by depth coverage (read count divided by gene length × 100) with gene length correction for rate loss [24].
Figure 1: Experimental workflow for comparative benchmarking of 16S rRNA and shotgun sequencing methodologies. The parallel processing paths enable direct performance comparison across multiple metrics including taxonomic resolution, diversity measures, and detection sensitivity.
Table 3: Key Research Reagent Solutions for Taxonomic Classification Studies
| Category | Product/Resource | Specifications | Application Notes |
|---|---|---|---|
| DNA Extraction | QIAamp Powerfecal DNA kit (Qiagen) | Standardized protocol for fecal samples | Optimal for both 16S and shotgun sequencing [8] |
| 16S Sequencing | LoopSeq 16S Microbiome Kit (Loop Genomics) | Full-length 16S (V1-V9) synthetic long-read technology | Improves species-level resolution compared to partial 16S [98] |
| Shotgun Library Prep | Nextera XT DNA Library Preparation Kit (Illumina) | Tagmentation-based library prep | Efficient fragmentation and adapter tagging [8] |
| Reference Database | SILVA SSU rRNA database (v138.1) | Curated 16S rRNA sequence database | Primary taxonomy assignment for 16S data [5] [98] |
| Reference Database | Meteor2 Microbial Gene Catalogues | Environment-specific gene catalogues | Supports 10 ecosystems; enables TFSP [24] |
| Bioinformatic Tool | DADA2 (v1.22.0) | Pipeline for ASV calling from 16S data | Implements error correction and chimera removal [5] |
| Bioinformatic Tool | Meteor2 | Tool for taxonomic, functional, and strain-level profiling | Leverages compact, environment-specific gene catalogues [24] |
| Classification Tool | MetaPhlAn4 | Marker-based taxonomic profiler | Part of bioBakery suite; species-level profiling [24] [100] |
Analysis and Interpretation Framework
Statistical Comparison and Validation
Diversity Metrics Calculation:
- Alpha Diversity: Calculate Observed OTUs/ASVs, Chao1, Shannon, and Simpson indices using consistent rarefaction depths across both methods [98]. Expect significantly higher alpha diversity estimates from shotgun sequencing, particularly in samples with adequate sequencing depth (>500,000 reads) [14].
- Beta Diversity: Perform Principal Coordinates Analysis (PCoA) based on Bray-Curtis dissimilarity matrices to visualize compositional differences between samples [8] [5]. Assess concordance between 16S and shotgun results through Procrustes analysis.
Differential Abundance Testing:
- Statistical Framework: Apply DESeq2 for identifying significantly differentially abundant taxa between experimental conditions [14]. Use adjusted p-value < 0.05 as significance threshold.
- Concordance Assessment: Compare fold changes of genera common to both sequencing strategies. In comparative studies, 93.3% of genera identified as significantly different by both methods show concordant fold changes between caeca and crop compartments [14].
Method-Specific Bias Evaluation:
- 16S Limitations: Identify taxa consistently underrepresented or missed by 16S sequencing, particularly low-abundance genera and species with high sequence similarity in targeted variable regions [14] [98].
- Shotgun Limitations: Monitor for database-dependent biases, where taxa without representative genomes in reference databases may be misclassified or missed entirely [5] [101].
Machine Learning for Predictive Modeling
Model Training and Validation:
- Feature Selection: Use genus-level or species-level abundance profiles as input features for predictive models of disease states [8] [5].
- Performance Comparison: Train separate random forest or logistic regression models on 16S and shotgun data from the same sample set [8] [5].
- Validation Approach: Employ cross-validation and independent test sets to assess prediction accuracy. Studies comparing pediatric ulcerative colitis classification reported area under the receiver operating characteristic curve (AUROC) of approximately 0.90 for both 16S and shotgun sequencing, despite differences in taxonomic resolution [8].
Signature Discovery:
- Microbial Signature Identification: Compare taxonomic features identified as important predictors in models trained on both data types [5]. Note that while both methods may identify overlapping signatures (e.g., Fusobacterium species in colorectal cancer), shotgun sequencing typically reveals additional discriminatory taxa [5].
Figure 2: Decision framework for evaluating sequencing method performance and selecting appropriate approaches based on research objectives. The framework incorporates multiple benchmarking metrics and acknowledges critical factors that influence methodological performance.
This benchmarking analysis demonstrates that both 16S rRNA and shotgun sequencing provide valuable insights into microbial community composition, but with distinct trade-offs that must be considered within specific research contexts. Shotgun metagenomics offers superior taxonomic resolution, enhanced detection of low-abundance taxa, and direct access to functional genetic elements, making it the preferred method for comprehensive microbiome characterization, particularly in hypothesis-driven research and therapeutic development [14] [24]. Conversely, 16S rRNA sequencing remains a cost-effective alternative for large-scale epidemiological studies or when budget constraints preclude deep shotgun sequencing, with the caveat that its limited resolution may obscure biologically significant patterns [8] [5].
The choice between these methodologies should be guided by specific research questions, sample types, and resource constraints. For drug development professionals seeking to identify novel therapeutic targets or biomarkers, shotgun sequencing provides the necessary resolution to detect subtle but potentially clinically relevant taxonomic and functional shifts. For longitudinal studies tracking microbial dynamics or large-scale population screenings, 16S sequencing may provide sufficient taxonomic information at a fraction of the cost. As sequencing technologies continue to evolve and costs decrease, hybrid approaches that combine shallow shotgun sequencing with targeted 16S analysis may offer optimal solutions for comprehensive microbiome study design.
Within the framework of shotgun metagenomic sequencing for microbiome functional analysis, the reconstruction of Metagenome-Assembled Genomes (MAGs) has emerged as a fundamental technique for studying uncultured microorganisms. MAGs allow researchers to access the genomic blueprints of the vast majority of microbes that resist laboratory cultivation, often referred to as microbial "dark matter" [102] [103]. The process involves assembling short sequencing reads into longer contiguous sequences (contigs) and subsequently binning these contigs into draft genomes that represent individual microbial populations within a complex community [102].
The critical importance of MAG validation lies in ensuring the accuracy and reliability of downstream biological interpretations. While MAGs provide unprecedented access to uncultivated lineages, the assembly and binning processes are inherently imperfect and can introduce errors, including contamination from other genomes, chimeric sequences, and fragmentation [104]. Validation against isolate genomes—derived from clonal cultures of microorganisms—provides the most stringent assessment of MAG quality, allowing researchers to quantify completeness, contamination, and taxonomic accuracy before making ecological inferences or proposing novel taxonomic groups [105]. This protocol details comprehensive methodologies for validating MAGs using isolate genomes, ensuring robust genome-resolved metagenomic research.
Background and Key Concepts
MAGs in Microbial Ecology
The transition from marker gene surveys (e.g., 16S rRNA sequencing) to whole-genome recovery via shotgun metagenomics represents a paradigm shift in microbial ecology [102]. While 16S sequencing provides community composition overviews, it lacks the resolution to distinguish closely related taxa and cannot directly access functional genetic elements [104] [41]. Shotgun metagenomics, by contrast, sequences DNA fragments randomly from all organisms in a sample, enabling not only higher taxonomic resolution but also functional profiling and genome reconstruction [1].
MAGs have dramatically expanded the known microbial tree of life, with recent analyses indicating that MAGs represent 48.54% of bacterial and 57.05% of archaeal diversity, compared to only 9.73% and 6.55% respectively for cultivated taxa [102]. This expansion has revealed novel taxa and metabolic pathways involved in critical biogeochemical cycles, including carbon, nitrogen, and sulfur transformations [102]. From a biomedical perspective, MAGs have enabled the identification of microbial constituents and functions associated with human health and disease states, such as liver conditions [106].
The Critical Need for Validation
Despite their utility, MAGs vary considerably in quality due to challenges in assembly and binning processes [104]. The complexity of microbial communities, varying genomic characteristics between taxa, and technical artifacts from sequencing and assembly algorithms can all introduce errors. Common issues include:
- Contamination: Presence of sequences from multiple organisms in a single MAG
- Fragmentation: Genomes split across multiple incomplete MAGs
- Chimerism: Artificial sequences created from parts of different genomes
- Taxonomic misassignment: Incorrect classification of the source organism
Validation against isolate genomes provides ground truth data to quantify these errors, ensuring that biological conclusions drawn from MAGs accurately reflect natural microbial systems rather than technical artifacts.
Validation Framework and Quality Standards
Established Quality Metrics
The scientific community has established standardized metrics for evaluating MAG quality through initiatives such as the Minimum Information about a Metagenome-Assembled Genome (MIMAG) standard [104]. These metrics provide objective criteria for assessing MAG quality:
Table 1: MAG Quality Standards Based on MIMAG Guidelines
| Quality Tier | Completeness | Contamination | Additional Criteria |
|---|---|---|---|
| High-quality | >90% | <5% | Presence of 16S, 23S, 5S rRNA genes and tRNA genes |
| Medium-quality | ≥50% | <10% | -- |
| Low-quality | <50% | >10% | -- |
High-quality MAGs (HMAGs) are essential for confident biological interpretation, particularly for proposing new taxa or making functional predictions [104]. In practice, a typical MAG database might contain HMAGs with mean completeness of 96.84% (± 2.81%) and mean contamination of 1.02% (± 1.09%), with genome sizes ranging from 0.52 to 12.26 Mb [104].
Validation Approaches
Multiple validation strategies exist, each with distinct advantages:
- Benchmarking with Mock Communities: Artificially constructed communities of known composition provide ground truth for validation [105]
- Comparison to Isolate Genomes: Direct comparison to sequenced isolates from similar environments
- Cross-pipeline Validation: Comparing results across multiple bioinformatics pipelines
- Taxonomic Consistency Checks: Ensuring phylogenetic markers yield consistent classifications
Each approach provides complementary evidence for MAG quality, with isolate genome comparison offering the most direct assessment when suitable reference genomes are available.
Experimental Protocol for MAG Validation
Sample Preparation and Sequencing
Critical Step: Sample selection and DNA extraction methods significantly impact downstream MAG quality and validation outcomes [102].
Materials:
- Sample Preservation: RNAlater or OMNIgene.GUT stabilization solutions for field sampling [102]
- DNA Extraction: PowerFecal DNA extraction kit or similar, optimized for high-molecular-weight DNA [106]
- Quality Assessment: NanoDrop spectrophotometer, Fragment Analyzer for library quality [106]
- Sequencing: Illumina NovaSeq for high-coverage sequencing (>20 million reads/sample recommended) [106]
Procedure:
- Sample Collection: Aseptically collect samples using sterile tools into DNA-free containers
- Preservation: Immediately freeze at -80°C or place in stabilization buffer
- DNA Extraction: Use mechanical lysis with bead beating followed by column-based purification
- Quality Control: Verify DNA integrity (A260/A280 ratio ~1.8-2.0), quantity (>1 ng/μL), and fragment size (>10 kb ideal)
- Library Preparation: Use Kapa Hyper Stranded kit or similar with appropriate adapters
- Sequencing: Perform paired-end sequencing (2×150 bp) on Illumina platform with sufficient depth (>20 million reads per sample)
Technical Note: For host-associated samples (e.g., human gut), remove host DNA contamination by aligning reads to the host genome (e.g., GRCh38 for human) using Bowtie2 and filtering mapped reads [106].
MAG Reconstruction and Binning
Materials:
- Computational Resources: High-performance computing cluster with adequate memory (≥128 GB RAM)
- Software Tools: BASALT [103], metaWRAP [104], VAMB [103], DAStool [103]
Procedure:
- Quality Control: Remove adapters and low-quality reads using FastQC and Trimmomatic
- Assembly: Perform metagenomic assembly using metaSPAdes or MEGAHIT
- Binning: Execute multiple binning tools (e.g., MaxBin2, CONCOCT, MetaBAT2)
- Bin Refinement: Use BASALT or metaWRAP to integrate and refine bins from multiple tools
- Quality Assessment: Evaluate completeness and contamination using CheckM
Technical Note: BASALT significantly improves binning quality by employing deep learning to identify core sequences, performing de-redundancy, decontamination, and fragment recovery, increasing high-quality genome yield by 2-fold compared to other tools [103].
Validation Against Isolate Genomes
Materials:
- Reference Genomes: NCBI RefSeq database, GTDB (Genome Taxonomy Database)
- Software Tools: CheckM, dRep, FastANI, GTDB-Tk
Procedure:
- Reference Selection: Identify closely related isolate genomes from public databases
- Quality Assessment: Calculate completeness and contamination using CheckM
- Taxonomic Classification: Assign taxonomy using GTDB-Tk
- Pairwise Comparison: Calculate Average Nucleotide Identity (ANI) using FastANI
- Phylogenetic Placement: Construct phylogenetic trees with single-copy marker genes
Validation Metrics:
- Completeness: Percentage of single-copy core genes present in MAG
- Contamination: Percentage of single-copy core genes present in multiple copies
- Strain Heterogeneity: Evidence for multiple strains within a single MAG
- ANI: Nucleotide identity between MAG and reference isolate (>95% for same species)
Table 2: Bioinformatics Tools for MAG Validation
| Tool | Primary Function | Validation Role | Key Features |
|---|---|---|---|
| CheckM | Quality assessment | Quantifies completeness/contamination | Uses lineage-specific marker genes |
| GTDB-Tk | Taxonomic classification | Validates taxonomic assignment | Standardized taxonomy based on reference tree |
| FastANI | Genome comparison | Measures similarity to isolates | Fast alignment-free ANI calculation |
| dRep | Genome de-replication | Identifies redundant MAGs | Clusters genomes based on ANI |
| BUSCO | Gene content assessment | Evaluates gene completeness | Universal single-copy orthologs |
Benchmarking and Performance Assessment
Comparative Pipeline Analysis
Recent benchmarking studies using mock communities provide critical insights into the performance of different bioinformatics pipelines for MAG recovery and analysis [105]. These assessments use metrics including sensitivity, false positive rates, and Aitchison distance (a compositional metric) to evaluate accuracy.
Table 3: Performance of Shotgun Metagenomic Pipelines Based on Mock Community Analysis
| Pipeline | Profiling Method | Strengths | Limitations | Sensitivity |
|---|---|---|---|---|
| bioBakery4 | Marker gene + MAG-based | Best overall accuracy metrics | Requires command line knowledge | High |
| JAMS | k-mer based (Kraken2) | High sensitivity | Resource-intensive assembly | Highest |
| WGSA2 | k-mer based (Kraken2) | High sensitivity | Optional assembly step | Highest |
| Woltka | Operational Genomic Units (OGUs) | Phylogenetic approach | No assembly performed | Moderate |
Key Finding: bioBakery4, which incorporates both marker genes and MAGs in its classification scheme (using species-genome bins or SGBs), demonstrates superior performance in most accuracy metrics [105]. This highlights the importance of integrated approaches that leverage both methods for optimal taxonomic profiling.
Impact of Sequencing Depth
Sequencing depth significantly influences both MAG completeness and the number of recoverable MAGs [104]. Deeper sequencing yields more high-quality MAGs, particularly for complex environments like soil or sediment. However, the relationship varies by sample type:
- Human gut samples: Progressive completeness declines with increasing sequencing depth
- Environmental samples: Completeness shows gradual increases or stable plateaus with depth
- All sample types: Number of recovered HMAGs increases with sequencing depth
These findings indicate that optimal sequencing strategies must consider microbial community complexity and research objectives when planning validation studies.
Research Toolkit
Table 4: Essential Research Reagents and Computational Tools for MAG Validation
| Category | Item | Function | Example/Alternative |
|---|---|---|---|
| Wet Lab | DNA Preservation Buffer | Stabilizes nucleic acids during storage/transport | RNAlater, OMNIgene.GUT |
| DNA Extraction Kit | Extracts high-molecular-weight DNA from samples | PowerFecal DNA Kit | |
| Library Prep Kit | Prepares sequencing libraries | Kapa Hyper Stranded Kit | |
| Quality Control Instruments | Assesses DNA and library quality | Fragment Analyzer, NanoDrop | |
| Bioinformatics | Binning Tools | Recovers MAGs from assembled contigs | BASALT, VAMB, DAStool, metaWRAP |
| Quality Assessment | Evaluates MAG completeness/contamination | CheckM, BUSCO | |
| Taxonomic Classification | Assigns taxonomy to MAGs | GTDB-Tk | |
| Genome Comparison | Compares MAGs to isolate genomes | FastANI, dRep | |
| Reference Databases | Provides isolate genomes for validation | GTDB, NCBI RefSeq, MAGdb |
Workflow Visualization
MAG Validation Workflow
Bioinformatics Validation Pipeline
Validating MAGs with isolate genomes represents a critical quality control step in shotgun metagenomic research, ensuring that genomic inferences accurately reflect biological reality. The integration of robust laboratory protocols, advanced computational tools like BASALT for binning refinement, and comprehensive benchmarking against isolate references provides a framework for generating high-quality MAGs suitable for downstream functional analysis and taxonomic discovery.
As MAG methodologies continue to evolve—with improvements in long-read sequencing, hybrid assembly approaches, and multi-omics integration—the validation principles outlined here will remain essential for maintaining scientific rigor in microbiome research. By adhering to these protocols, researchers can confidently exploit MAGs to illuminate microbial dark matter, advancing our understanding of microbiome function in health, disease, and environmental sustainability.
Shotgun metagenomic sequencing has revolutionized microbiome research by enabling comprehensive analysis of the functional potential of microbial communities. Moving beyond taxonomic census, this approach allows researchers to quantify the abundance of functional genes and metabolic pathways encoded within a microbiome, revealing its capacity to influence and respond to its environment [37]. Correlating this functional gene abundance with concurrent environmental metadata is a powerful strategy to generate testable hypotheses about microbial community function and its impact on host or ecosystem health [27] [107]. This case study outlines a standardized protocol for conducting such an integrated analysis, framed within a broader research thesis on shotgun metagenomic sequencing for microbiome functional analysis. It is designed for an audience of researchers, scientists, and drug development professionals seeking to implement robust metagenomic correlation studies.
Experimental Design and Workflow
A typical project correlating functional gene abundance with environmental metadata follows a multi-stage workflow, from sample collection through to integrated data analysis. The key steps are summarized in the diagram below.
Sample Collection and Metadata Acquisition
The initial phase involves the simultaneous collection of biological samples and their associated environmental metadata.
- Sample Type: The choice of sample matrix (e.g., stool, soil, water) dictates subsequent processing protocols. For human gut studies, rectal swabs or fecal samples are commonly used [27].
- Metadata Collection: A rigorous and standardized approach to capturing environmental data is crucial. This includes:
- Clinical/Physiological Data: Disease status, patient demographics, diet, medication use [27].
- Environmental Parameters: For ecological studies, this includes pH, temperature, nutrient concentrations, and gas flux measurements (e.g., N2O) [108].
- Sample Metadata: Time of collection, storage conditions, and processing delays.
- Preservation: Immediate stabilization of samples is critical to preserve the authentic microbial community and nucleic acid integrity. Swab or soil samples should be snap-frozen (e.g., in liquid nitrogen) and stored at -80°C prior to DNA extraction [108] [27].
Detailed Experimental Protocols
DNA Extraction and Shotgun Metagenomic Sequencing
The goal of this protocol is to obtain high-quality, high-molecular-weight DNA that is representative of the entire microbial community, suitable for shotgun sequencing.
Materials & Reagents:
- MP-soil FastDNA Spin Kit for Soil (#6560-200, MP Biomedicals) or equivalent for efficient cell lysis and DNA purification from complex matrices [27].
- Agarose Gel Electrophoresis System (e.g., 1% agarose gel) for qualitative DNA assessment.
- Spectrophotometer (e.g., NanoDrop 2000, Thermo Fisher Scientific) and Fluorometer (e.g., TBS-380) for quantifying DNA concentration and purity [27].
Procedure:
- Cell Lysis: Follow manufacturer's instructions for the DNA extraction kit. Mechanical lysis (e.g., bead beating) is often necessary to break down robust microbial cell walls, especially for soil and stool samples [37].
- DNA Purification: Bind and wash DNA on the provided spin columns to remove contaminants like humic acids (soil) or host compounds (stool) that can inhibit downstream enzymatic reactions [37].
- Quality Control (QC):
- Assess DNA purity using a spectrophotometer (A260/A280 ratio ~1.8-2.0 is ideal).
- Quantify DNA concentration using a fluorometer for accuracy.
- Verify DNA integrity via agarose gel electrophoresis to confirm high molecular weight and absence of excessive degradation [27].
- Library Preparation and Sequencing:
- Convert qualified DNA into a sequencing library using an Illumina-compatible kit (e.g., Illumina HiSeq 4000 platform) [27].
- Perform paired-end sequencing (e.g., 2x150 bp) to a minimum depth of 10 million reads per sample for adequate functional coverage, though deeper sequencing (e.g., 10-14 Gb) is recommended for high-resolution analysis [27].
Bioinformatic Processing and Functional Profiling
This protocol processes raw sequencing reads into quantified functional gene and pathway abundances.
Software & Databases:
- Fastp (v0.23.0+) for quality control and adapter trimming [27].
- Bowtie2 (v2.5.4+) or BWA (v0.7.17+) for removing host-associated reads by aligning to a host genome (e.g., human GRCh38) [24] [27].
- Meteor2 for comprehensive taxonomic, functional, and strain-level profiling (TFSP) [24].
- HUMAnN3 or HUMAnN4 as an alternative for functional profiling from metagenomic reads [24] [33].
- KEGG Database for functional orthology (KO) and pathway annotation (e.g., KEGG Modules, Gut Metabolic Modules) [24] [27].
- DIAMOND (v2.0.13+) for fast alignment of reads against protein databases like KEGG [27].
- R Package 'vegan' (v4.0.5+) for diversity analysis and statistical modeling [27].
Procedure:
- Quality Control & Preprocessing:
- Host DNA Depletion:
- Align reads to the host reference genome using
BWA. Discard reads that align to the host to focus analysis on the microbial fraction [27].
- Align reads to the host reference genome using
- Functional Profiling:
- Option A (Using Meteor2):
- Align quality-filtered reads to a specific microbial gene catalogue (e.g., human intestinal catalogue) using
bowtie2[24]. - Quantify gene abundances using Meteor2's default "shared" counting mode, which proportionally distribates multi-mapping reads [24].
- Annotate genes against KEGG Orthology (KO), Carbohydrate-Active Enzymes (CAZymes), and Antibiotic Resistance Genes (ARGs) using Meteor2's integrated pipelines [24].
- Normalize gene counts using depth coverage or FPKM (Fragments Per Kilobase Million) [24].
- Align quality-filtered reads to a specific microbial gene catalogue (e.g., human intestinal catalogue) using
- Option B (Using HUMAnN3/4):
- Option A (Using Meteor2):
- Pathway Abundance Computation:
- Aggregate the abundances of genes annotated with the same KO identifier to estimate the abundance of higher-order functional modules like KEGG modules or Gut Metabolic Modules (GMMs) [24].
Statistical Integration and Correlation Analysis
This protocol details the statistical methods for identifying significant associations between functional features and environmental metadata.
Procedure:
- Data Normalization and Transformation:
- Normalize functional abundance tables (e.g., KO abundances, pathway abundances) to relative abundance or transform them using methods like Centered Log-Ratio (CLR) for compositional data analysis.
- Standardize environmental metadata to a common scale (z-scores) if variables are on different units.
- Diversity Analysis:
- Differential Abundance Testing:
- Correlation Analysis:
- Perform multivariate ordination analyses, such as Redundancy Analysis (RDA) or Canonical Correspondence Analysis (CCA), to model the relationship between the entire functional profile and a set of environmental variables.
- Calculate pairwise correlation coefficients (e.g., Spearman's rank correlation) between individual functional features and individual metadata variables. Correct for multiple hypothesis testing using methods like Benjamini-Hochberg False Discovery Rate (FDR).
Key Reagents and Computational Tools
Table 1: Essential Research Reagent Solutions and Computational Tools
| Item Name | Type | Function/Purpose | Example/Catalog Number |
|---|---|---|---|
| MP-soil FastDNA Spin Kit | Reagent Kit | Efficient DNA extraction from complex samples like soil and stool | MP Biomedicals #6560-200 [27] |
| Illumina HiSeq 4000 | Sequencing Platform | High-throughput shotgun metagenomic sequencing | Illumina HiSeq 4000 [27] |
| PacBio HiFi Sequencing | Sequencing Platform | Long-read sequencing for improved assembly and strain resolution | PacBio Sequel II/IIe Systems [33] |
| KEGG Database | Bioinformatics Database | Functional annotation of genes and reconstruction of metabolic pathways | KEGG v94.2+ [27] [24] |
| Meteor2 | Software Pipeline | Integrated taxonomic, functional, and strain-level profiling | Meteor2 [24] |
| HUMAnN3/4 | Software Pipeline | Profiling the abundance of microbial metabolic pathways and gene families | HUMAnN3 / HUMAnN4 [24] [33] |
| Fastp | Software Tool | Quality control and adapter trimming of raw sequencing reads | fastp (v0.23.0+) [27] |
| Bowtie2 | Software Tool | Aligning sequencing reads to reference genomes or gene catalogues | Bowtie2 (v2.5.4+) [24] [27] |
Data Presentation and Analysis
The following table illustrates the type of results generated from a differential analysis of functional pathways, as one might find in a study of gut microbiome changes during disease recovery.
Table 2: Example Functional Pathway Analysis During Recovery from Acute Pancreatitis (AP). Data derived from a shotgun metagenomic study of rectal swabs from AP patients [27].
| Functional Pathway (KEGG Level 2/3) | Trend in Mild AP (MAP) Recovery | Trend in Severe AP (SAP) Recovery | Potential Biological Interpretation |
|---|---|---|---|
| Short-Chain Fatty Acid (SCFA) Synthesis | Increasing | Decreasing/Stable | Restoration of beneficial microbes; improved gut barrier integrity [27] |
| Lipopolysaccharide (LPS) Biosynthesis | Decreasing | Increasing | Reduction in pro-inflammatory potential in MAP; persistent inflammation risk in SAP [27] |
| Antibiotic Resistance Genes (ARGs) | Decreasing | Increasing | Reduction in pathobionts in MAP; selection for resistant organisms in SAP [24] [27] |
| Nitrogen Metabolism | Varies | Varies | Altered urea processing and ammonia levels, potentially linked to patient's nitrogen balance [108] |
| Carbohydrate-Active Enzymes (CAZymes) | Increasing (specific types) | Decreasing | Recovery of microbial capacity to metabolize dietary fibers in MAP [24] |
The logical flow from raw data to biological insight in a correlation analysis can be visualized as follows.
This application note provides a detailed framework for conducting a robust case study to correlate functional gene abundance with environmental metadata. By integrating rigorous laboratory protocols for shotgun metagenomics with advanced bioinformatic processing and statistical modeling, researchers can move from mere observations of microbial membership to actionable insights about community function. The standardized workflows and tools outlined here, such as the Meteor2 pipeline and the integration of KEGG annotations, empower scientists to generate reproducible data, formulate testable hypotheses about microbiome-environment interactions, and ultimately identify functional targets for therapeutic intervention or biomarker development in human health, agriculture, and environmental science.
Cross-Platform and Cross-Protocol Reproducibility in Metagenomic Studies
Shotgun metagenomic sequencing has revolutionized our ability to decipher the taxonomic composition and functional potential of complex microbial communities. For research in drug development and clinical diagnostics, achieving reproducible results across different laboratories and sequencing platforms is paramount. However, the multifaceted nature of metagenomic workflows—encompassing DNA extraction, library preparation, sequencing technology, and bioinformatics analysis—introduces significant variability that can compromise the comparability of findings across studies [109]. This application note synthesizes recent evidence to outline standardized protocols and analytical frameworks that enhance reproducibility, providing researchers with practical strategies for robust microbiome functional analysis.
Results
Impact of Wet-Lab Methodologies on Reproducibility
DNA Extraction Kits
The initial DNA extraction step critically influences downstream results, with significant variability observed in DNA yield, quality, and microbial representation among commercial kits. A comprehensive cross-comparison of four DNA isolation kits revealed substantial differences in performance when applied to canine stool samples, a validated model for human gut microbiome research [109].
Table 1: Performance Comparison of DNA Extraction Kits
| Kit Manufacturer | Average DNA Yield | DNA Quality (Degradation) | Host DNA Ratio | Hands-on Time | Inter-replicate Reproducibility |
|---|---|---|---|---|---|
| Qiagen (Q) | Lowest | Highest degradation | Significantly higher | Moderate | Low (failed in 6/6 canine samples) |
| Invitrogen (I) | Moderate | Suitable for LRS | Low | Moderate | Highest variance |
| Macherey-Nagel (MN) | Highest | Suitable for LRS | Low | Moderate | High |
| Zymo Research (Z) | High (with half sample volume) | High-quality, HMW DNA suitable for LRS WGS | Low | Most extensive | Most consistent, minimal variation |
The Zymo Research Quick-DNA HMW MagBead Kit produced the most consistent results with minimal variation among replicates and yielded high-molecular-weight (HMW) DNA essential for long-read sequencing (LRS) applications. In contrast, the Qiagen kit consistently produced the lowest yield and highest degradation, with significantly higher host DNA contamination [109].
Library Preparation and Sequencing Platforms
The choice between short-read sequencing (SRS) and long-read sequencing (LRS) platforms, as well as specific library preparation methods, introduces another layer of technical variability. In the same evaluation, the Illumina DNA Prep library preparation method emerged as particularly effective for high-quality microbial diversity analysis [109].
Cross-platform comparisons between BGISEQ-500 and Illumina platforms (HiSeq 2000 and HiSeq 4000) demonstrated high intra-platform reproducibility, with sequence and library replicates showing Spearman correlation coefficients >0.91 at the gene level and >0.97 at the species level. However, cross-platform comparisons revealed that 2.02%–3.25% of genes exhibited significant differences in relative abundance, with a bias toward genes with higher GC content being enriched on Illumina HiSeq platforms [110].
Table 2: Sequencing Platform Performance Metrics
| Platform | High-Quality Reads (%) | Q30 Bases (%) | IGC Mapping Rate (%) | Unique Mapping Rate (%) | Intra-platform Spearman Correlation (Gene Level) |
|---|---|---|---|---|---|
| BGISEQ-500 | 96.06% | 90.56% | 77.77% | 63.27% | >0.91 |
| HiSeq 2000 | Not specified | Not specified | Comparable | Comparable | >0.91 (Illumina platforms) |
Bioinformatics Strategies for Enhanced Reproducibility
Bioinformatics analysis introduces substantial variability in metagenomic studies, with tool selection significantly impacting taxonomic and functional profiles. A cross-comparison of bioinformatics approaches revealed that the effectiveness of specific pipelines is often sample-dependent, making it challenging to identify a universally optimal approach [109].
To address this challenge, the study developed minitax, a software tool designed to provide consistent results across platforms and methodologies. Following alignment with minimap2 to reference databases, minitax determines the most probable taxonomy for each read based on mapping qualities (MAPQ values) and CIGAR strings, offering a uniform analytical approach for diverse data types [109].
For visualization and exploratory analysis, tools like Metaviz enable interactive statistical and visual analysis of metagenomic data. Metaviz implements a FacetZoom navigation control that allows dynamic aggregation of abundance counts across taxonomic hierarchies, facilitating reproducible exploration of complex datasets [111].
Experimental Protocols
DNA Extraction Protocol for High-Molecular-Weight Microbial DNA
Based on: Zymo Research Quick-DNA HMW MagBead Kit [109]
Reagents and Equipment
- Zymo Research Quick-DNA HMW MagBead Kit
- Covaris E220 ultrasonicator (or equivalent)
- Axygen AxyPrep Mag PCR Clean-Up Kit
- TE Buffer
- Bead beater for mechanical lysis
Procedure
- Sample Preparation: Aliquot 200 mg of frozen stool sample into a bead-beating tube.
- Cell Lysis: Add 800 µL of HMW Lysis Buffer and perform bead-beating for 10 minutes to ensure efficient disruption of Gram-positive bacterial cells.
- Enzymatic Treatment: Incubate with proteinase K (20 mg/mL) at 55°C for 2 hours with gentle agitation.
- DNA Binding: Transfer supernatant to magnetic beads and incubate for 15 minutes with periodic mixing.
- Washing: Perform two washes with HMW Wash Buffer, ensuring complete removal of supernatant.
- Elution: Elute DNA in 50 µL of TE Buffer pre-heated to 55°C.
- Quality Assessment: Verify DNA integrity via pulsed-field gel electrophoresis and quantify using fluorometric methods.
Cross-Platform Sequencing Validation Protocol
Based on: BGISEQ-500 and Illumina HiSeq Comparison [110]
Reagents and Equipment
- BGISEQ-500 platform (BGI)
- Illumina HiSeq 2000/4000 platform
- Covaris LE220 ultrasonicator
- AMPure XP beads (Beckman Coulter)
- MetaHIT protocol reagents for DNA extraction
Library Preparation and Sequencing
- DNA Fragmentation: Fragment 500 ng of input DNA ultrasonically to 300-700 bp fragments using Covaris E220.
- Library Construction: Perform end-repairing, A-tailing, and adapter ligation using barcoded adapters.
- PCR Amplification: Amplify libraries with 8-10 cycles to minimize bias.
- Quality Control: Assess library size distribution and quantity using Bioanalyzer and qPCR.
- Sequencing: Process libraries on both BGISEQ-500 (SE100 mode) and Illumina HiSeq platforms (100 bp paired-end).
- Data Processing: Apply overall accuracy (OA) quality control method to filter reads, retaining only those with >96% high-quality bases.
Reproducible Bioinformatics Analysis Protocol
Based on: minitax Implementation [109]
Software Requirements
- minitax (custom tool for taxonomic profiling)
- minimap2 aligner
- Reference genome collection or 16S database
- Python 3.8+ with Biopython, pandas
Analysis Workflow
- Data Preprocessing: Convert raw sequencing reads to FASTQ format and perform quality control.
- Alignment: Map reads to reference database using minimap2 with default parameters.
- Taxonomic Assignment: For each read, identify the best alignment and determine taxonomy using MAPQ values and CIGAR string parsing.
- Abundance Profiling: Generate count tables for taxonomic groups at desired hierarchical levels.
- Cross-Platform Normalization: Apply variance-stabilizing transformation to account for platform-specific technical effects.
The Scientist's Toolkit
Table 3: Essential Research Reagents and Tools for Reproducible Metagenomic Studies
| Category | Product/Kit | Specific Function | Performance Notes |
|---|---|---|---|
| DNA Extraction | Zymo Research Quick-DNA HMW MagBead Kit | High-molecular-weight DNA extraction with bead-beating | Highest consistency, minimal host DNA, suitable for LRS |
| Library Preparation | Illumina DNA Prep | Metagenomic whole genome sequencing library construction | Most effective for high-quality microbial diversity analysis |
| Sequencing Platforms | BGISEQ-500 | High-throughput sequencing using cPAS technology | High intra-platform reproducibility, comparable to Illumina |
| Sequencing Platforms | Illumina HiSeq 4000 | Short-read sequencing with reversible terminators | Industry standard, but shows GC bias compared to BGISEQ-500 |
| Bioinformatics | minitax | Taxonomic profiling across platforms | Provides consistent results across sequencing methodologies |
| Bioinformatics | Metaviz | Interactive visualization of taxonomic hierarchies | Enables dynamic exploration and reproducible visual analysis |
| Quality Control | Overall Accuracy (OA) Control | Quality adjustment for sequencing data | Retains 96.06% of raw reads as high-quality, with 90.56% Q30 bases |
Visualizations
Experimental Workflow for Reproducible Metagenomics
Bioinformatics Reproducibility Pipeline
Discussion
Achieving reproducibility in metagenomic studies requires careful consideration of each step in the workflow, from sample collection to data analysis. The evidence presented demonstrates that methodological choices at each stage significantly impact results, potentially confounding cross-study comparisons and biological interpretations.
The identification of optimal practices for specific stages—such as the Zymo Research kit for DNA extraction, Illumina DNA Prep for library construction, and minitax for bioinformatics analysis—provides a foundation for standardizing methodologies [109]. However, the sample-dependent effectiveness of these combinations underscores the importance of employing multiple approaches or validating methods for specific sample types.
For drug development professionals, these reproducibility challenges have direct implications on biomarker discovery and therapeutic development. The growing application of microbiome profiling in precision medicine, particularly in oncology for early cancer detection and predicting immunotherapy response, heightens the need for standardized, reproducible methods [112]. Emerging approaches such as analyzing cell-free microbial DNA (cf-mbDNA) for liquid biopsy applications offer promising alternatives but will require similar rigorous validation of cross-platform reproducibility [112].
Future directions should focus on developing community-standardized protocols for specific sample types and research questions, along with continued refinement of bioinformatics tools that normalize platform-specific biases. The integration of interactive visualization tools like Metaviz with robust analytical pipelines will further enhance reproducible exploration and interpretation of complex metagenomic datasets [111].
Conclusion
Shotgun metagenomic sequencing has revolutionized microbiome science by providing unparalleled access to the functional genetic potential of microbial communities, moving beyond taxonomic census to mechanistic understanding. This comprehensive overview demonstrates its critical role in pharmaceutical development, from tracking antimicrobial resistance to discovering novel therapeutics and understanding drug-microbiome interactions. Future directions point toward integrating multi-omics data, developing standardized analytical frameworks, and translating microbial functional insights into personalized medical interventions and advanced biotechnological applications. As computational methods advance and sequencing costs decrease, shotgun metagenomics is poised to become an indispensable tool in clinical diagnostics, therapeutic development, and understanding host-microbe relationships in health and disease.
References
- 1. Shotgun Metagenomic Sequencing [https://www.illumina.com/areas-of-interest/microbiology/microbial-sequencing-methods/shotgun-metagenomic-sequencing.html]
- 2. Shotgun Metagenomic Sequencing Guide [https://blog.microbiomeinsights.com/shotgun-metagenomic-sequencing-guide]
- 3. The Power of Shotgun Metagenomics in Environmental ... [https://www.cd-genomics.com/resource-power-shotgun-metagenomics-environmental-researches.html]
- 4. Unravel Your Metagenomic Sample - Shotgun Sequencing ... [https://cegat.com/unravel-your-metagenomic-sample-shotgun-sequencing-vs-16s-sequencing/]
- 5. Comparison between 16S rRNA and shotgun sequencing in ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-024-10621-7]
- 6. Comprehensive evaluation of shotgun metagenomics ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9939430/]
- 7. An introduction to the analysis of shotgun metagenomic data [https://pmc.ncbi.nlm.nih.gov/articles/PMC4059276/]
- 8. 16S rRNA and metagenomic shotgun sequencing data ... [https://www.nature.com/articles/s41598-022-07995-7]
- 9. Sequential co-assembly reduces computational resources ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12049710/]
- 10. Next-generation sequencing: insights to advance clinical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8970668/]
- 11. 16S rRNA Sequencing Guide [https://blog.microbiomeinsights.com/16s-rrna-sequencing-guide]
- 12. 16S vs Metagenomics: What's the Difference? [https://www.cosmosid.com/blog/16s-vs-metagenomics-whats-the-difference/]
- 13. 16S rRNA Gene Sequencing vs. Shotgun Metagenomic ... [https://blog.microbiomeinsights.com/16s-rrna-sequencing-vs-shotgun-metagenomic-sequencing]
- 14. Comparison between 16S rRNA and shotgun sequencing ... [https://www.nature.com/articles/s41598-021-82726-y]
- 15. The Human Microbiome and Understanding the 16S rRNA ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5535273/]
- 16. Unculturable bacteria—the uncharacterized organisms that ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1279316/]
- 17. Unculturable Bacteria and Their Significance in Contemporary ... [https://microbewiki.kenyon.edu/index.php/Unculturable_Bacteria_and_Their_Significance_in_Contemporary_Medicine]
- 18. Uncultured Microorganisms and Their Functions in the ... [https://www.mdpi.com/2304-8158/12/14/2691]
- 19. Unculturable Microbes → Term [https://lifestyle.sustainability-directory.com/term/unculturable-microbes/]
- 20. Metagenomics for studying unculturable microorganisms [https://pmc.ncbi.nlm.nih.gov/articles/PMC1273625/]
- 21. Shotgun Metagenomic Sequencing Service [https://www.zymoresearch.com/pages/shotgun-metagenomic-sequencing-service?srsltid=AfmBOorR1Sm9t5mf6bMT-M7_Es3XxBx4CdQ4vCjHfWMRce0lz1STboHT]
- 22. How to Determine the Role of the Microbiome in Drug ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7333656/]
- 23. Metagenomic approaches in microbial ecology: an update ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7641418/]
- 24. Accurate profiling of microbial communities for shotgun ... [https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-025-02249-w]
- 25. EasyMetagenome: A userâ€friendly and flexible pipeline for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11865343/]
- 26. MEDUSA: A Pipeline for Sensitive Taxonomic ... [https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2022.814437/full]
- 27. Metagenomics reveals functional profiles of gut microbiota ... [https://www.nature.com/articles/s41598-025-05127-5]
- 28. borenstein-lab/MetaLAFFA: A bioinformatics pipeline for ... [https://github.com/borenstein-lab/MetaLAFFA]
- 29. Session 9 – Graphics and Colors | Bioinformatics Workshop ... [https://corytophanes.github.io/BIO_BIT_Bioinformatics_209/graphics-and-colors.html]
- 30. Ten simple rules to colorize biological data visualization - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7561171/]
- 31. Mock community taxonomic performance of publicly... classification [https://www.nature.com/articles/s41597-023-02877-7?error=cookies_not_supported&code=2f3ee756-d231-4936-a3a4-3c4df805e1d1]
- 32. Guide Metagenomics Sequencing [https://genohub.com/shotgun-metagenomics-sequencing/]
- 33. 2025 Microbiome SMRT Grant Awardees [https://www.pacb.com/blog/2025-microbiome-smrt-grant-awardees/]
- 34. Integrating taxonomic signals from MAGs and contigs ... [https://www.nature.com/articles/s41467-024-47155-1]
- 35. Chapter 4 Bin annotation [https://www.earthhologenome.org/bioinformatics/bin-annotation.html]
- 36. Shotgun metagenomics of the vaginal microbiome in ... [https://www.nature.com/articles/s41598-025-23737-x]
- 37. Metagenomics - a guide from sampling to data analysis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3351745/]
- 38. Assessment of protocols for characterization of the human ... [https://pubmed.ncbi.nlm.nih.gov/41196055/]
- 39. Shotgun Metagenomics Sequencing [https://www.completegenomics.com/areas-of-interest/microbiology/shotgun-metagenomics-sequencing/]
- 40. Functional analysis of microbiome by shotgun ... [https://bio-protocol.org/exchange/minidetail?id=10512455&type=30]
- 41. A practical guide to amplicon and metagenomic analysis of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8106563/]
- 42. Data Processing and Visualization for Metagenomics [https://gwu-libraries.github.io/metagenomics-analysis/aio/index.html]
- 43. Review From bag-of-genes to bag-of-genomes: metabolic ... [https://www.sciencedirect.com/science/article/pii/S2001037020303172]
- 44. Evaluating metagenomics tools for genome binning with real ... [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-03667-3]
- 45. Binning sequences using very sparse labels within a ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2383919/]
- 46. MG-RAST user manual [https://help.mg-rast.org/user_manual.html]
- 47. NCBI Prokaryotic Genome Annotation Pipeline - NIH [https://www.ncbi.nlm.nih.gov/refseq/annotation_prok/]
- 48. Comprehensive multi omics explore the microbial function ... [https://www.nature.com/articles/s41538-025-00575-9]
- 49. Shotgun Metagenomics Reveals Antibiotic Resistome ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1626799/full]
- 50. Microbial metabolic enzymes, pathways and ... [https://www.sciencedirect.com/science/article/pii/S0043135425001435]
- 51. Integrated analysis of blood microbiome and metabolites ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-025-06165-3]
- 52. MAID Shotgun Metagenomics Course | Centre for Genomic ... [https://www.liverpool.ac.uk/genomic-research/training-and-events/maid-shotgun-metagenomics-course/]
- 53. Advances in functional analysis of the microbiome [https://pubmed.ncbi.nlm.nih.gov/39895076/]
- 54. Plasmids, a Molecular Cornerstone of Antimicrobial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12440250/]
- 55. Mobilizable plasmids drive the spread of antimicrobial ... [https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-023-01260-w]
- 56. AMRViz enables seamless genomics analysis and ... [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-024-05792-9]
- 57. Accurate profiling of microbial communities for shotgun ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12590682/]
- 58. The best practice for microbiome analysis using R - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10599642/]
- 59. Data-Driven Approaches in Antimicrobial Resistance [https://pmc.ncbi.nlm.nih.gov/articles/PMC11590962/]
- 60. WHO releases new reports on new tests and treatments in ... [https://www.who.int/news/item/02-10-2025-who-releases-new-reports-on-new-tests-and-treatments-in-development-for-bacterial-infections]
- 61. Antimicrobial resistance: Not enough antibacterials in the ... [https://www.soci.org/news/2025/10/antimicrobial-resistance-not-enough-antibacterials-in-the-pipeline-says-who]
- 62. Building microbial communities to improve antimicrobial ... [https://www.nature.com/articles/s44259-025-00115-1]
- 63. Introduction to Shotgun Metagenomics, from Sampling ... [https://www.cd-genomics.com/introduction-to-shotgun-metagenomics-from-sampling-to-data-analysis.html]
- 64. Microbial genome mining for accelerated natural products ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4070288/]
- 65. Four therapeutic frontiers opened up by strain-level ... [https://www.ddw-online.com/four-therapeutic-frontiers-opened-up-by-strain-level-microbiome-sequencing-37596-202510/]
- 66. Genome mining-driven discovery of enzymes catalyzing ... [https://www.nature.com/articles/s41429-025-00881-0]
- 67. Drug-microbiota interactions: an emerging priority for ... [https://www.nature.com/articles/s41392-023-01619-w]
- 68. Gut Microbiome Interactions with Drug Metabolism, Efficacy ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5718288/]
- 69. Introduction to Microbiome Analysis [https://training.galaxyproject.org/training-material/topics/microbiome/tutorials/introduction/slides-plain.html]
- 70. Gut microbiome metagenomics in clinical practice [https://pmc.ncbi.nlm.nih.gov/articles/PMC12562794/]
- 71. Benefits and challenges of host depletion methods in ... [https://www.nature.com/articles/s41522-025-00762-2]
- 72. A metagenomic DNA sequencing assay that is robust ... [https://www.nature.com/articles/s41467-022-31654-0]
- 73. Simple statistical identification and removal of contaminant ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6298009/]
- 74. Guidelines for preventing and reporting contamination in ... [https://www.nature.com/articles/s41564-025-02035-2]
- 75. Reduction of PCR-amplifiable DNA by ethylene oxide ... [https://www.sciencedirect.com/science/article/abs/pii/S1872497314001240]
- 76. Shotgun metagenomics sequencing | IDT [https://www.idtdna.com/pages/applications/infectious-disease/shotgun-metagenomics-for-infectious-diseases]
- 77. Shotgun Metagenomic Sequencing: Determining Depth [https://blog.microbiomeinsights.com/shotgun-metagenomic-sequencing-determining-depth]
- 78. Shallow shotgun sequencing reduces technical variation in ... [https://www.nature.com/articles/s41598-023-33489-1]
- 79. Shallow Shotgun Sequencing [https://microbiomeinsights.com/shallow-shotgun-sequencing/]
- 80. Amplicon vs Shotgun Metagenomic Sequencing [https://genesmart.vn/en/amplicon-sequencing-vs-shotgun-metagenomics-choosing-the-right-approach-for-microbiome-research/]
- 81. A Shotgun Metagenomics Benchmarking Case Study [https://seqwell.com/tradeoffs-of-miniaturization-a-shotgun-metagenomics-benchmarking-case-study/]
- 82. Metagenome-Assembled Genomes (MAGs): Advances, ... [https://www.mdpi.com/2076-2607/13/5/985]
- 83. Metagenomic co-assembly uncovers mobile antibiotic ... [https://www.nature.com/articles/s43247-025-02381-3]
- 84. Metagenomics Unveils Viral Biodiversity [https://virologyresearchservices.com/2025/10/28/metagenomics-unveils-viral-biodiversity/]
- 85. Computational Metagenomics: State of the Art [https://www.mdpi.com/1422-0067/26/18/9206]
- 86. Ten simple rules for investigating (meta)genomic data from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9731419/]
- 87. a scalable and resource-efficient workflow for multi-strategy ... [https://pubmed.ncbi.nlm.nih.gov/41036626/]
- 88. Data Processing and Visualization for Metagenomics: Figures [https://carpentries-lab.github.io/metagenomics-analysis/figures/index.html]
- 89. NMDC Data Management Best Practices [https://microbiomedata.org/data-management/]
- 90. Tools for Analysis of the Microbiome - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7598837/]
- 91. Shotgun and Hi-C Sequencing Datasets for Binning Wheat ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11873117/]
- 92. Benchmarking metagenomic binning tools on real datasets ... [https://www.nature.com/articles/s41467-025-57957-6]
- 93. Protocol for refining metagenomic binning with BinSPreader [https://www.sciencedirect.com/science/article/pii/S2666166723003842]
- 94. How Do You Sequence a Microbiome? 5 Steps Explained [https://www.cosmosid.com/blog/how-do-you-sequence-a-microbiome-5-steps-explained/]
- 95. Calculating fast differential genome coverages among ... [https://www.nature.com/articles/s42003-025-09007-6]
- 96. Microbial functional pathways based on metatranscriptomic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10847690/]
- 97. Comparative Analysis of 16S rRNA Gene and ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2021.670336/full]
- 98. The effect of taxonomic classification by full-length 16S ... [https://www.nature.com/articles/s41598-020-80826-9]
- 99. Refining microbiome diversity analysis by concatenating and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11993755/]
- 100. Benchmarking bacterial taxonomic classification using ... [https://www.nature.com/articles/s41597-024-03672-8]
- 101. Benchmarking Metagenomics Tools for Taxonomic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6716367/]
- 102. Metagenome-Assembled Genomes (MAGs): Advances ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12114606/]
- 103. ä½™ç‚团队在《Nature Communications》ç‰æœŸåˆŠå‘表了挖掘 ... [https://news.pkusz.edu.cn/info/1003/8190.htm]
- 104. MAGdb: a comprehensive high quality MAGs repository for ... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03711-6]
- 105. Mock community taxonomic classification performance of ... [https://www.nature.com/articles/s41597-023-02877-7]
- 106. Deep sequencing-derived Metagenome Assembled ... [https://www.nature.com/articles/s41597-024-04153-8]
- 107. Functional prediction based on 16S rRNA metagenome ... [https://www.nature.com/articles/s41598-024-69538-6]
- 108. Development of an Environmental Functional Gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2976231/]
- 109. Cross-comparison of gut metagenomic profiling strategies [https://www.nature.com/articles/s42003-024-07158-6]
- 110. Assessment of the cPAS-based BGISEQ-500 platform for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5848809/]
- 111. Metaviz: interactive statistical and visual analysis of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5887897/]
- 112. The microbiome in precision medicine [https://www.ddw-online.com/the-microbiome-in-precision-medicine-24877-202307/]